Analyses ‘ceasing to publish’
Last compiled on februari, 2024
This lab journal replicates the analyses for ‘ceasing to publish’.
Custom functions
package.check: Check if packages are installed (and install if not) in R (source).
fpackage.check <- function(packages) {
lapply(packages, FUN = function(x) {
if (!require(x, character.only = TRUE)) {
install.packages(x, dependencies = TRUE)
library(x, character.only = TRUE)
}
})
}
fsave <- function(x, file, location="./data/processed/") {
datename <- substr(gsub("[:-]", "", Sys.time()), 1,8)
totalname <- paste(location, datename, file, sep="")
save(x, file = totalname)
}Packages
survival: general package to create life tables and survival models, needed for flexsurvflexsurv: fitting flexible survival regression modelsboot: bootstrapping for SEs of AMEstidyverse: for data manipulationggplot2: for creating figures 5-7ggpubr: for combining two figures in one (plot 5)kableExtra: formatting tables
packages = c("survival", "flexsurv", "boot", "tidyverse", "ggplot2", "ggpubr", "kableExtra")
fpackage.check(packages)## Loading required package: survival## Loading required package: flexsurv## Loading required package: boot##
## Attaching package: 'boot'## The following object is masked from 'package:survival':
##
## aml## Loading required package: kableExtra##
## Attaching package: 'kableExtra'## The following object is masked from 'package:dplyr':
##
## group_rowsInput
We use two processed datasets.
- df_stopping.rda:
person-period file containing publications and all relevant variables
for the survival models, time window for inactivity is 3 years
- For construction of this dataset see Dependent Variables : Starting and Stopping
to Publish
- name of dataset:
df_ppf3
- For construction of this dataset see Dependent Variables : Starting and Stopping
to Publish
- df_stopping5.rda:
person-period file containing publications and all relevant variables
for the survival models, time-window for inactivity is years
- For construction of this dataset see Dependent Variables : Starting and Stopping
to Publish
- name of dataset:
df_ppf5
- For construction of this dataset see Dependent Variables : Starting and Stopping
to Publish
load(file = "./data/processed/df_stopping.rda")
load(file = "data/processed/df_stopping5.rda")Checking distributions
Kaplan-Meier curve
KM1 <- survfit(Surv(time-1, time, inactive) ~ 1, conf.lower='usual', data = df_ppf3)
KM2 <- survfit(Surv(time-1, time, inactive) ~ gender, conf.lower='usual', data = df_ppf3)
KM3 <- survfit(Surv(time-1, time, inactive) ~ ethnicity2, conf.lower='usual', data = df_ppf3)
plot(KM1)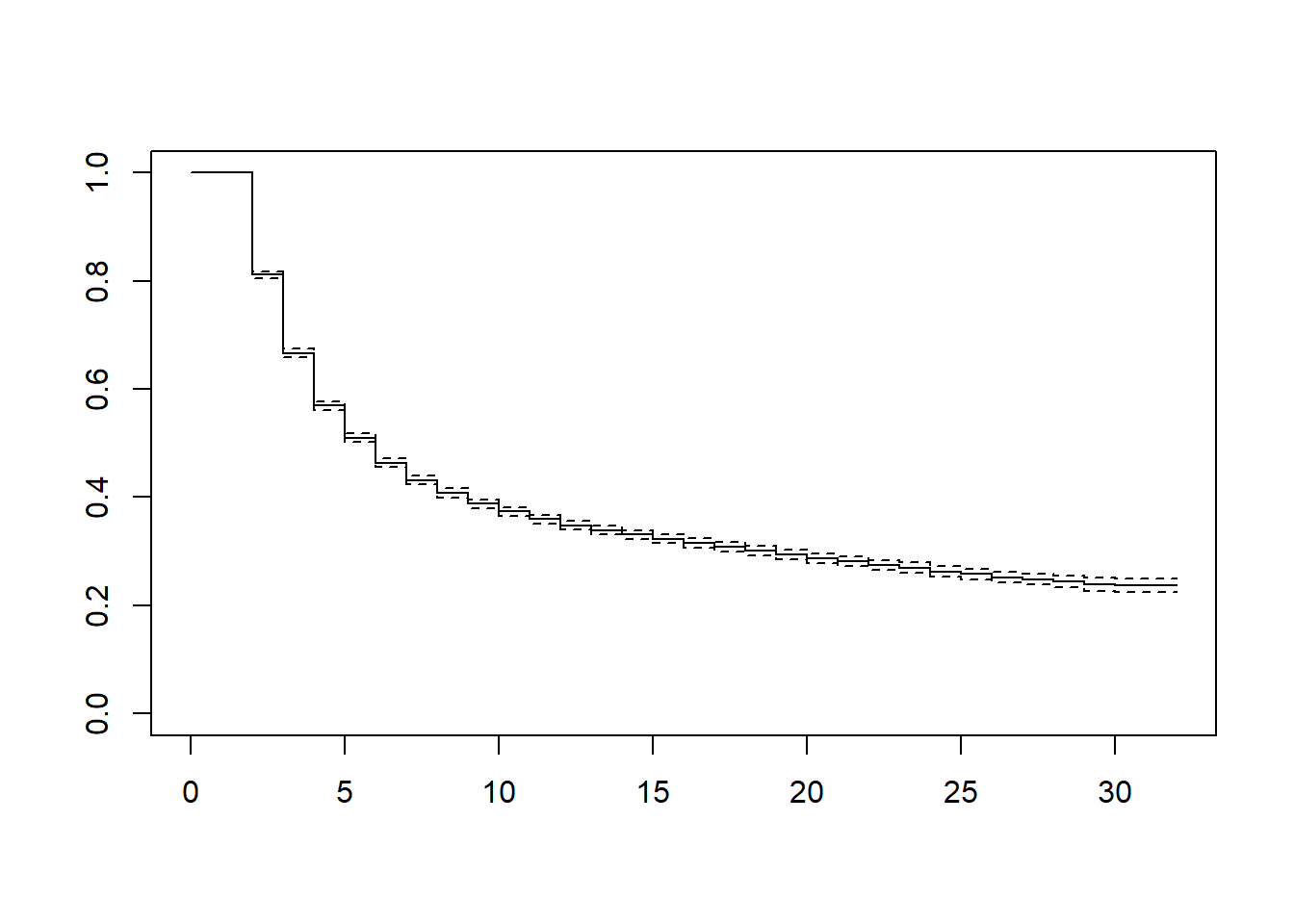
plot(KM2)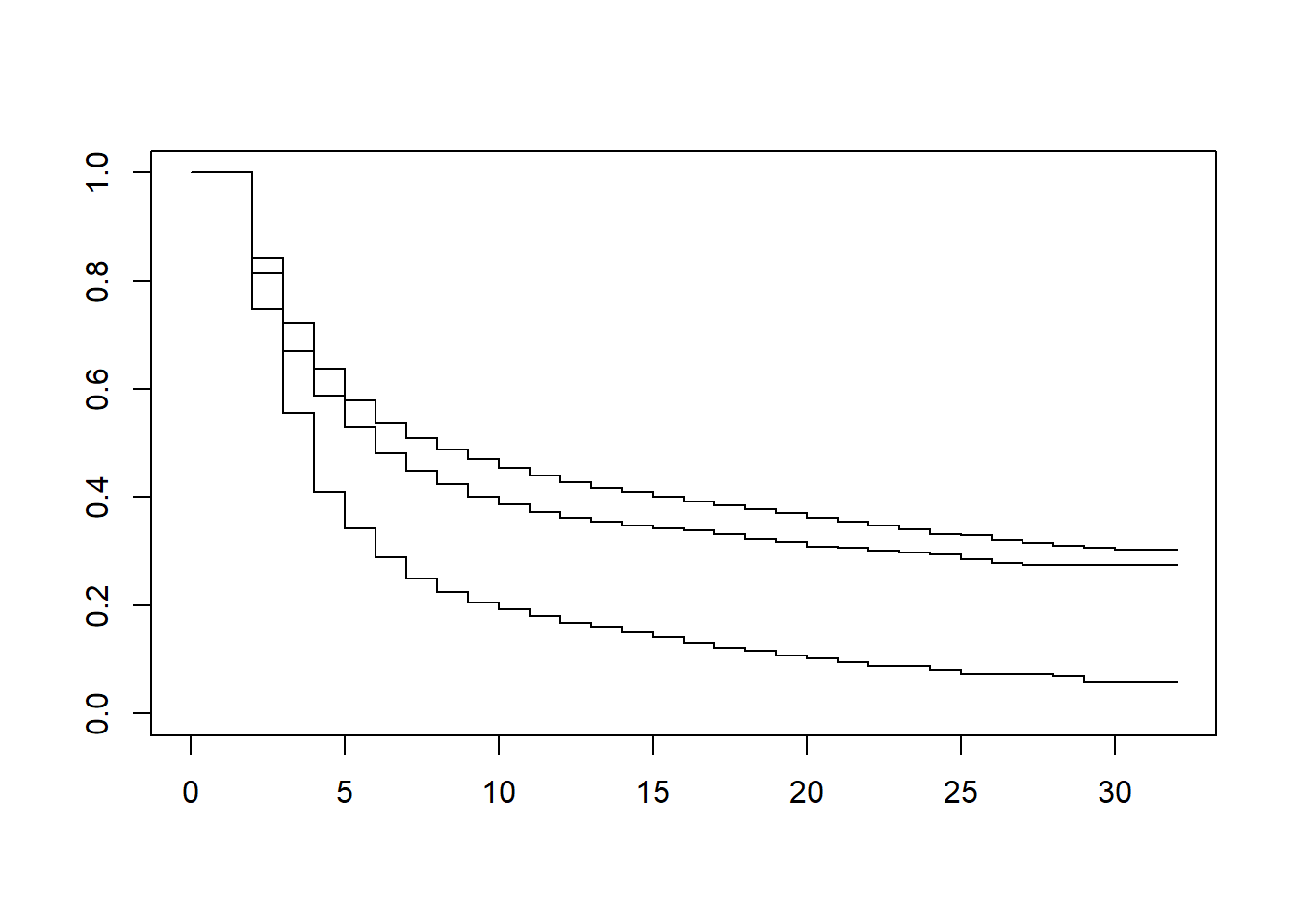
plot(KM3)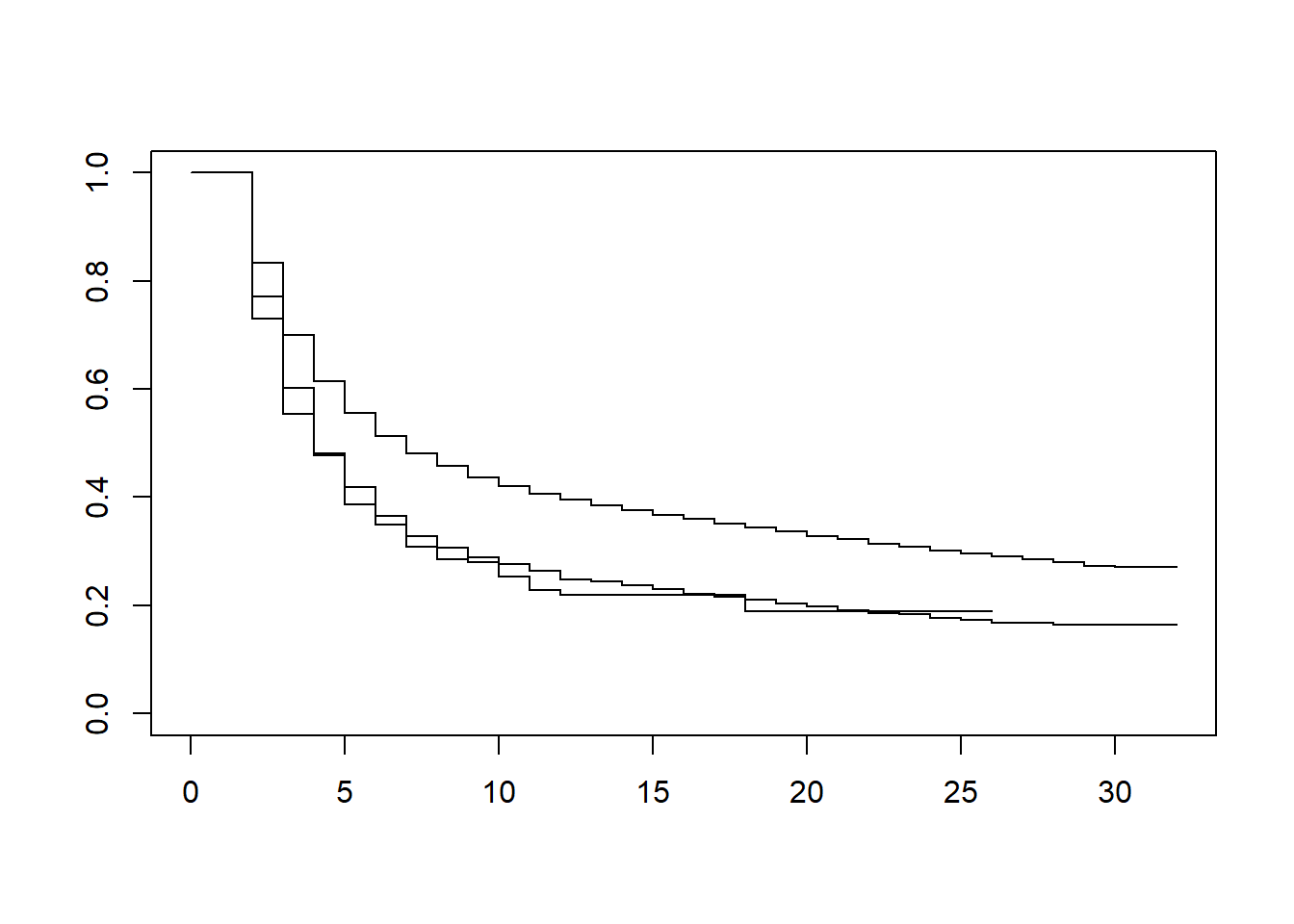
#MC1 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "gengamma") # commented: no convergence
#MC2 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "genf") # commented: no convergence
MC3 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "weibull")
MC4 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "gamma")
MC5 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "exp")
MC6 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "llogis")
MC7 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "gompertz")
MC8 <- flexsurvreg(Surv((time -1), time, inactive) ~ 1, data = df_ppf3, dist = "lognormal")
testdist <- AIC(MC3, MC4, MC5, MC6, MC7, MC8)
testdist[order(testdist$AIC),]Model 1: Gender only
M1 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender , data = df_ppf3, dist = "lognormal")M1## Call:
## flexsurvreg(formula = Surv((time - 1), time, inactive) ~ gender,
## data = df_ppf3, dist = "lognormal")
##
## Estimates:
## data mean est L95% U95% se exp(est) L95% U95%
## meanlog NA 2.19181 2.16380 2.21983 0.01429 NA NA NA
## sdlog NA 1.06590 1.05003 1.08202 0.00816 NA NA NA
## genderwomen 0.32431 -0.19212 -0.23351 -0.15073 0.02112 0.82521 0.79175 0.86008
## gendermissing 0.15973 -0.62256 -0.66863 -0.57649 0.02351 0.53657 0.51241 0.56187
##
## N = 118372, Events: 9951, Censored: 108421
## Total time at risk: 118372
## Log-likelihood = -32564.76, df = 4
## AIC = 65137.52P-values
## meanlog sdlog genderwomen gendermissing
## 0 0 0 0Model 2: Ethnicity only
M2 <- flexsurvreg(Surv((time -1), time, inactive) ~ ethnicity2 , data = df_ppf3, dist = "lognormal")M2 ## Call:
## flexsurvreg(formula = Surv((time - 1), time, inactive) ~ ethnicity2,
## data = df_ppf3, dist = "lognormal")
##
## Estimates:
## data mean est L95% U95% se exp(est) L95% U95%
## meanlog NA 2.11350 2.09063 2.13636 0.01167 NA NA NA
## sdlog NA 1.07443 1.05840 1.09070 0.00824 NA NA NA
## ethnicity2minority 0.01282 -0.45499 -0.58923 -0.32075 0.06849 0.63445 0.55475 0.72561
## ethnicity2other 0.24807 -0.37859 -0.41738 -0.33980 0.01979 0.68483 0.65877 0.71191
##
## N = 118372, Events: 9951, Censored: 108421
## Total time at risk: 118372
## Log-likelihood = -32721.55, df = 4
## AIC = 65451.1P-values
## meanlog sdlog ethnicity2minority ethnicity2other
## 0 0 0 0Model 3: Gender and ethnicity, controls
M3 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s, data = df_ppf3, dist = "lognormal")M3## Call:
## flexsurvreg(formula = Surv((time - 1), time, inactive) ~ gender +
## ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s, data = df_ppf3,
## dist = "lognormal")
##
## Estimates:
## data mean est L95% U95% se
## meanlog NA 2.254134 2.172277 2.335991 0.041765
## sdlog NA 0.847069 0.834590 0.859736 0.006415
## genderwomen 0.324308 -0.084489 -0.118045 -0.050932 0.017121
## gendermissing 0.159734 -0.436816 -0.475709 -0.397922 0.019844
## ethnicity2minority 0.012824 -0.119125 -0.224714 -0.013536 0.053873
## ethnicity2other 0.248065 -0.101543 -0.133789 -0.069298 0.016452
## uniLU 0.016947 -0.054197 -0.173059 0.064666 0.060645
## uniRU 0.211317 -0.205849 -0.284479 -0.127219 0.040118
## uniRUG 0.040846 0.608518 0.457734 0.759303 0.076932
## uniTUD 0.004131 0.367652 0.163204 0.572100 0.104312
## uniTUE 0.023840 0.503403 0.342315 0.664492 0.082190
## uniTI 0.010873 -0.435195 -0.578198 -0.292192 0.072962
## uniUM 0.096568 -0.023207 -0.109659 0.063245 0.044109
## uniUT 0.080543 -0.605701 -0.691606 -0.519796 0.043830
## uniUU 0.090376 -0.181588 -0.265715 -0.097462 0.042922
## uniUvA 0.121828 -0.049879 -0.136390 0.036633 0.044139
## uniVU 0.090748 -0.048208 -0.137242 0.040825 0.045426
## uniWUR 0.158940 -0.000619 -0.085835 0.084596 0.043478
## field2Physical and Mathematical Sciences 0.116016 -0.201981 -0.248623 -0.155340 0.023797
## field2Social and Behavioral Sciences 0.145904 0.193687 0.146858 0.240516 0.023893
## field2Engineering 0.077620 -0.119362 -0.176828 -0.061897 0.029320
## field2Agricultural Sciences 0.085899 0.048752 -0.010329 0.107833 0.030144
## field2Humanities 0.041429 0.293647 0.209210 0.378085 0.043081
## field2missing 0.160798 0.290978 0.243449 0.338508 0.024250
## phd_cohort 15.109840 -0.033922 -0.036236 -0.031609 0.001180
## npubs_prev_s 0.479051 1.500639 1.434418 1.566861 0.033787
## exp(est) L95% U95%
## meanlog NA NA NA
## sdlog NA NA NA
## genderwomen 0.918982 0.888656 0.950343
## gendermissing 0.646090 0.621444 0.671714
## ethnicity2minority 0.887697 0.798745 0.986556
## ethnicity2other 0.903442 0.874775 0.933049
## uniLU 0.947246 0.841088 1.066802
## uniRU 0.813956 0.752406 0.880540
## uniRUG 1.837707 1.580489 2.136786
## uniTUD 1.444340 1.177277 1.771985
## uniTUE 1.654342 1.408203 1.943503
## uniTI 0.647138 0.560908 0.746625
## uniUM 0.977060 0.896139 1.065288
## uniUT 0.545692 0.500771 0.594642
## uniUU 0.833945 0.766658 0.907137
## uniUvA 0.951345 0.872502 1.037312
## uniVU 0.952935 0.871759 1.041670
## uniWUR 0.999381 0.917746 1.088278
## field2Physical and Mathematical Sciences 0.817110 0.779874 0.856124
## field2Social and Behavioral Sciences 1.213716 1.158189 1.271905
## field2Engineering 0.887486 0.837924 0.939980
## field2Agricultural Sciences 1.049960 0.989724 1.113862
## field2Humanities 1.341311 1.232703 1.459487
## field2missing 1.337736 1.275641 1.402853
## phd_cohort 0.966647 0.964413 0.968886
## npubs_prev_s 4.484555 4.197201 4.791582
##
## N = 118372, Events: 9951, Censored: 108421
## Total time at risk: 118372
## Log-likelihood = -30292, df = 26
## AIC = 60636P-values
## meanlog sdlog
## 0.0000 0.0000
## genderwomen gendermissing
## 0.0000 0.0000
## ethnicity2minority ethnicity2other
## 0.0270 0.0000
## uniLU uniRU
## 0.3715 0.0000
## uniRUG uniTUD
## 0.0000 0.0004
## uniTUE uniTI
## 0.0000 0.0000
## uniUM uniUT
## 0.5988 0.0000
## uniUU uniUvA
## 0.0000 0.2585
## uniVU uniWUR
## 0.2886 0.9886
## field2Physical and Mathematical Sciences field2Social and Behavioral Sciences
## 0.0000 0.0000
## field2Engineering field2Agricultural Sciences
## 0.0000 0.1058
## field2Humanities field2missing
## 0.0000 0.0000
## phd_cohort npubs_prev_s
## 0.0000 0.0000Average marginal effects (AMEs)
bootFunc <- function(data, i) {
df <- data[i,] #bootstrap datasets
M <- flexsurv::flexsurvreg(survival::Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s, data = df, dist = "lognormal")
suppressWarnings({
dfmaj <- dfmin <- dfmen <- dfwomen <- df
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity2 <- "minority" # one dataset all minority
dfmaj$ethnicity2 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df$gender))
dfmin$ethnicity2 <- factor(dfmin$ethnicity2, levels=levels(df$ethnicity2))
dfmaj$ethnicity2 <- factor(dfmaj$ethnicity2, levels=levels(df$ethnicity2))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(M, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(M, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(M, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(M, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
})
c(AME_gender, AME_ethnicity)
#save results
}
b3 <- boot(df_ppf3, bootFunc, R = 999, parallel="snow", ncpus=10)
fsave(b3, file = "boot3.rda", location = "./results/stopping/")original <- b3$t0
bias <- colMeans(b3$t) - b3$t0
se <- apply(b3$t, 2, sd)
boot.df3 <- data.frame(original=original, bias=bias, se=se)
row.names(boot.df3) <- c("AME_gender", "AME_ethnicity")
boot.df3$t <- (boot.df3$original / boot.df3$se)
round(boot.df3, 5)## original bias se t
## AME_gender -1.65428 -0.00432 0.31677 -5.22231
## AME_ethnicity -2.18740 0.04751 0.84055 -2.60236Model 4: Gender + ethnicity + cohort x gender + cohort x ethnicity + controls
M4 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_ppf3, dist = "lognormal")M4## Call:
## flexsurvreg(formula = Surv((time - 1), time, inactive) ~ gender +
## ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort *
## gender + phd_cohort * ethnicity2, data = df_ppf3, dist = "lognormal")
##
## Estimates:
## data mean est L95% U95% se
## meanlog NA 2.241756 2.152108 2.331405 0.045740
## sdlog NA 0.845462 0.833004 0.858107 0.006404
## genderwomen 0.324308 0.021328 -0.074926 0.117582 0.049110
## gendermissing 0.159734 -0.656435 -0.759920 -0.552949 0.052800
## ethnicity2minority 0.012824 -0.138562 -0.533041 0.255916 0.201268
## ethnicity2other 0.248065 -0.020699 -0.118039 0.076640 0.049664
## uniLU 0.016947 -0.036353 -0.155113 0.082408 0.060593
## uniRU 0.211317 -0.195520 -0.274085 -0.116955 0.040085
## uniRUG 0.040846 0.606243 0.455623 0.756863 0.076848
## uniTUD 0.004131 0.392737 0.188261 0.597213 0.104326
## uniTUE 0.023840 0.515919 0.354808 0.677030 0.082201
## uniTI 0.010873 -0.416125 -0.559084 -0.273166 0.072940
## uniUM 0.096568 -0.004576 -0.091134 0.081982 0.044163
## uniUT 0.080543 -0.584547 -0.670719 -0.498374 0.043966
## uniUU 0.090376 -0.164626 -0.248936 -0.080315 0.043016
## uniUvA 0.121828 -0.034374 -0.121055 0.052307 0.044226
## uniVU 0.090748 -0.033359 -0.122402 0.055684 0.045431
## uniWUR 0.158940 0.010904 -0.074224 0.096032 0.043434
## field2Physical and Mathematical Sciences 0.116016 -0.202237 -0.248801 -0.155672 0.023758
## field2Social and Behavioral Sciences 0.145904 0.190841 0.144093 0.237589 0.023852
## field2Engineering 0.077620 -0.118354 -0.175720 -0.060987 0.029269
## field2Agricultural Sciences 0.085899 0.044306 -0.014673 0.103284 0.030092
## field2Humanities 0.041429 0.293140 0.208847 0.377434 0.043008
## field2missing 0.160798 0.291224 0.243740 0.338708 0.024227
## phd_cohort 15.109840 -0.033855 -0.037081 -0.030628 0.001646
## npubs_prev_s 0.479051 1.496518 1.430389 1.562648 0.033740
## genderwomen:phd_cohort 5.472333 -0.005237 -0.010027 -0.000446 0.002444
## gendermissing:phd_cohort 2.550983 0.012432 0.006944 0.017920 0.002800
## ethnicity2minority:phd_cohort 0.242718 0.000462 -0.017883 0.018808 0.009360
## ethnicity2other:phd_cohort 4.313131 -0.004433 -0.009240 0.000374 0.002453
## exp(est) L95% U95%
## meanlog NA NA NA
## sdlog NA NA NA
## genderwomen 1.021557 0.927813 1.124774
## gendermissing 0.518697 0.467704 0.575251
## ethnicity2minority 0.870609 0.586818 1.291645
## ethnicity2other 0.979513 0.888661 1.079654
## uniLU 0.964300 0.856318 1.085899
## uniRU 0.822407 0.760267 0.889625
## uniRUG 1.833530 1.577155 2.131579
## uniTUD 1.481029 1.207148 1.817048
## uniTUE 1.675177 1.425907 1.968024
## uniTI 0.659598 0.571733 0.760966
## uniUM 0.995435 0.912896 1.085436
## uniUT 0.557359 0.511341 0.607517
## uniUU 0.848211 0.779630 0.922825
## uniUvA 0.966210 0.885985 1.053699
## uniVU 0.967191 0.884793 1.057263
## uniWUR 1.010964 0.928463 1.100795
## field2Physical and Mathematical Sciences 0.816902 0.779735 0.855840
## field2Social and Behavioral Sciences 1.210267 1.154992 1.268188
## field2Engineering 0.888382 0.838853 0.940835
## field2Agricultural Sciences 1.045302 0.985434 1.108807
## field2Humanities 1.340631 1.232256 1.458537
## field2missing 1.338065 1.276013 1.403134
## phd_cohort 0.966712 0.963598 0.969836
## npubs_prev_s 4.466113 4.180324 4.771440
## genderwomen:phd_cohort 0.994777 0.990023 0.999554
## gendermissing:phd_cohort 1.012510 1.006968 1.018082
## ethnicity2minority:phd_cohort 1.000462 0.982276 1.018986
## ethnicity2other:phd_cohort 0.995577 0.990802 1.000374
##
## N = 118372, Events: 9951, Censored: 108421
## Total time at risk: 118372
## Log-likelihood = -30274.33, df = 30
## AIC = 60608.66P-values
## meanlog sdlog
## 0.0000 0.0000
## genderwomen gendermissing
## 0.6641 0.0000
## ethnicity2minority ethnicity2other
## 0.4912 0.6768
## uniLU uniRU
## 0.5485 0.0000
## uniRUG uniTUD
## 0.0000 0.0002
## uniTUE uniTI
## 0.0000 0.0000
## uniUM uniUT
## 0.9175 0.0000
## uniUU uniUvA
## 0.0001 0.4370
## uniVU uniWUR
## 0.4628 0.8018
## field2Physical and Mathematical Sciences field2Social and Behavioral Sciences
## 0.0000 0.0000
## field2Engineering field2Agricultural Sciences
## 0.0001 0.1409
## field2Humanities field2missing
## 0.0000 0.0000
## phd_cohort npubs_prev_s
## 0.0000 0.0000
## genderwomen:phd_cohort gendermissing:phd_cohort
## 0.0322 0.0000
## ethnicity2minority:phd_cohort ethnicity2other:phd_cohort
## 0.9606 0.0707Average marginal effects (AMEs)
bootFunc <- function(data, i) {
df <- data[i,] #bootstrap datasets
M <- flexsurv::flexsurvreg(survival::Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df, dist = "lognormal")
suppressWarnings({
dfmaj <- dfmin <- dfmen <- dfwomen <- df
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity2 <- "minority" # one dataset all minority
dfmaj$ethnicity2 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df$gender))
dfmin$ethnicity2 <- factor(dfmin$ethnicity2, levels=levels(df$ethnicity2))
dfmaj$ethnicity2 <- factor(dfmaj$ethnicity2, levels=levels(df$ethnicity2))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(M, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(M, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(M, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(M, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
# cohort interactions
s <- 0.1
# copying our data into separate dataframes
dfwomencp <- dfwomencm <- dfmencp <- dfmencm <- df
dfmincp <- dfmincm <- dfmajcp <- dfmajcm <- df
# assigning gender to each of the dataframes
dfwomencp$gender <- dfwomencm$gender <- "women"
dfmencp$gender <- dfmencm$gender <- "men"
dfwomencp$gender <- factor(dfwomencp$gender, levels=levels(df$gender))
dfwomencm$gender <- factor(dfwomencm$gender, levels=levels(df$gender))
dfmencp$gender <- factor(dfmencp$gender, levels=levels(df$gender))
dfmencm$gender <- factor(dfmencm$gender, levels=levels(df$gender))
# assigning ethnicity to the dataframes
dfmincp$ethnicity2 <- dfmincm$ethnicity2 <- "minority"
dfmajcp$ethnicity2 <- dfmajcm$ethnicity2 <- "majority"
dfmincp$ethnicity2 <- factor(dfmincp$ethnicity2, levels=levels(df$ethnicity2))
dfmincm$ethnicity2 <- factor(dfmincm$ethnicity2, levels=levels(df$ethnicity2))
dfmajcp$ethnicity2 <- factor(dfmajcp$ethnicity2, levels=levels(df$ethnicity2))
dfmajcm$ethnicity2 <- factor(dfmajcm$ethnicity2, levels=levels(df$ethnicity2))
# adding/subtracting small changes PhD cohort
dfwomencp$phd_cohort <- dfmencp$phd_cohort <- df$phd_cohort + s
dfwomencm$phd_cohort <- dfmencm$phd_cohort <- df$phd_cohort - s
dfmajcp$phd_cohort <- dfmincp$phd_cohort <- df$phd_cohort + s
dfmajcm$phd_cohort <- dfmincm$phd_cohort <- df$phd_cohort - s
# calculating predicted probabilities
pwp <- as.numeric(unlist(predict(M, type="response", newdata=dfwomencp)))
pwm <- as.numeric(unlist(predict(M, type="response", newdata=dfwomencm)))
pmp <- as.numeric(unlist(predict(M, type="response", newdata=dfmencp)))
pmm <- as.numeric(unlist(predict(M, type="response", newdata=dfmencm)))
pnp <- as.numeric(unlist(predict(M, type="response", newdata=dfmincp)))
pnm <- as.numeric(unlist(predict(M, type="response", newdata=dfmincm)))
pjp <- as.numeric(unlist(predict(M, type="response", newdata=dfmajcp)))
pjm <- as.numeric(unlist(predict(M, type="response", newdata=dfmajcm)))
# marginal effects
AME_gendercoh <- mean(((pwp - pmp) - (pwm - pmm)) / (2*s))
AME_ethnicitycoh <- mean(((pnp - pjp) - (pnm - pjm)) / (2*s))
})
c(AME_gender, AME_ethnicity, AME_gendercoh, AME_ethnicitycoh)
#save results
}
b4 <- boot(df_ppf3, bootFunc, R = 999, parallel="snow", ncpus=10)
fsave(b4, file = "boot4.rda", location = "./results/stopping/")original <- b4$t0
bias <- colMeans(b4$t) - b4$t0
se <- apply(b4$t, 2, sd)
boot.df4 <- data.frame(original=original, bias=bias, se=se)
row.names(boot.df4) <- c("AME_gender", "AME_ethnicity", "AME_gender-coh", "AME_ethnicity-coh")
boot.df4$t <- (boot.df4$original / boot.df4$se)
round(boot.df4, 5)## original bias se t
## AME_gender -0.78358 0.00137 0.51239 -1.52926
## AME_ethnicity -2.43144 0.15562 1.67483 -1.45175
## AME_gender-coh -0.07530 -0.00054 0.06601 -1.14074
## AME_ethnicity-coh 0.09160 -0.01590 0.20778 0.44085Table 4
columns <- c(rep(c("B", "C.I."), 4))
rows <- c("<strong>Shape</strong></p>", "<strong>Scale</strong></p>", "Gender: ref=men" ,"Women", "Missing gender", "Ethnicity: ref=majority","Minority", "Other", "University: ref=Erasmus University", "Leiden University", "Radboud University", "University of Groningen", "Delft University of Technology", "Eindhoven University of Technology", "Tilburg University", "Maastricht University", "University of Twente", "Utrecht University", "University of Amsterdam", "Vrije Universiteit Amsterdam", "Wageningen University and Research Centre", "Field: ref=Biological and Health Sciences", "Physical and mathematical sciences", "Social and behavioral sciences", "Engineering", "Agricultural sciences", "Humanities", "Missing field", "<strong>PhD cohort</strong></p>", "<strong>Previous publications</strong></p>", "Cohort interactions","PhD cohort * women", "PhD cohort * missing gender", "PhD cohort * ethnic minority", "PhD cohort * other ethnicity", "<strong>AIC", "<strong>N")
t4 <- data.frame(matrix(nrow=length(rows), ncol=length(columns)))
colnames(t4) <- columns
rownames(t4) <- rows
# Model 1
t4[c(1,2,4,5),1] <- format(round(as.numeric(M1$coef), 2), nsmall=2) # estimates
t4[c(1,2),2] <- paste0("[", format(round(log(M1$res[c(1,2),2]), 2), nsmall=2), " , ", format(round(log(M1$res[c(1,2),3]), 2), nsmall=2), "]") # CI for shape and scale
t4[c(4,5),2] <- paste0("[", format(round(M1$res[c(3,4),2], 2), nsmall=2), " , ", format(round(M1$res[c(3,4),3], 3), nsmall=2), "]") # CI for other covariates
# Model 2
t4[c(1,2,7,8),3] <- format(round(M2$coef, 2), nsmall=2)
t4[c(1,2),4] <- paste0("[", format(round(log(M2$res[c(1,2),2]), 2), nsmall=2), " , ", format(round(log(M2$res[c(1,2),3]), 2), nsmall=2), "]")
t4[c(7,8),4] <- paste0("[", format(round(M2$res[c(3,4),2], 2), nsmall=2), " , ", format(round(M2$res[c(3,4),3], 2), nsmall=2), "]")
# Model 3
t4[c(1,2,4,5,7,8,10:21,23:30),5] <- format(round(M3$coef, 2), nsmall=2)
#t4[c(1,2), 6] <- paste("[", round(log(M3$res[c(1,2),2]), 3), ",", round(log(M3$res[c(1,2),3]), 3), "]")
t4[c(1,2,4,5,7,8,10:21,23:30), 6] <- paste0("[", format(round(M3$res[c(1,2,3:26),2], 2), nsmall=2), " , ", format(round(M3$res[c(1,2,3:26),3], 2), nsmall=2), "]")
# Model 4
t4[c(1,2,4,5,7,8,10:21,23:30,32:35),7] <- format(round(M4$coef, 2), nsmall=2)
#t4[c(1,2), 8] <- paste("[", round(log(M4$res[c(1:2),2]), 3), ",", round(log(M4$res[c(1:2),3]), 3), "]")
t4[c(1,2,4,5,7,8,10:21,23:30,32:35), 8] <- paste0("[", format(round(M4$res[c(1,2,3:30),2], 2), nsmall=2), " , ", format(round(M4$res[c(1,2,3:30),3], 2), nsmall=2), "]")
# AIC
t4[36,1] <- round(M1$AIC, 0)
t4[36,3] <- round(M2$AIC, 0)
t4[36,5] <- round(M3$AIC, 0)
t4[36,7] <- round(M4$AIC, 0)
# sample size
t4[37, c(1,3,5,7)] <- rep(nrow(df_ppf3[!duplicated(df_ppf3$id),]), 4)
t4[is.na(t4)] <- ""t4 %>%
kable(format = 'html', caption = '<b>Table 4.</b> Log-normal regression analyses on stopping to publish', escape=FALSE) %>%
add_header_above(.,c(' '=1, 'Model 1'=2, 'Model 2'=2, 'Model 3'=2, 'Model 4'=2), escape = FALSE, bold = TRUE) %>%
row_spec(row=c(3,6,9,22,31), bold=T) %>%
kable_classic(full_width = F, html_font = "Cambria") %>%
kable_styling(font_size = 11) -> table4
table4| B | C.I. | B | C.I. | B | C.I. | B | C.I. | |
|---|---|---|---|---|---|---|---|---|
| Shape | 2.19 | [0.77 , 0.80] | 2.11 | [0.74 , 0.76] | 2.25 | [ 2.17 , 2.34] | 2.24 | [ 2.15 , 2.33] |
| Scale | 0.06 | [0.05 , 0.08] | 0.07 | [0.06 , 0.09] | -0.17 | [ 0.83 , 0.86] | -0.17 | [ 0.83 , 0.86] |
| Gender: ref=men | ||||||||
| Women | -0.19 | [-0.23 , -0.151] | -0.08 | [-0.12 , -0.05] | 0.02 | [-0.07 , 0.12] | ||
| Missing gender | -0.62 | [-0.67 , -0.576] | -0.44 | [-0.48 , -0.40] | -0.66 | [-0.76 , -0.55] | ||
| Ethnicity: ref=majority | ||||||||
| Minority | -0.45 | [-0.59 , -0.32] | -0.12 | [-0.22 , -0.01] | -0.14 | [-0.53 , 0.26] | ||
| Other | -0.38 | [-0.42 , -0.34] | -0.10 | [-0.13 , -0.07] | -0.02 | [-0.12 , 0.08] | ||
| University: ref=Erasmus University | ||||||||
| Leiden University | -0.05 | [-0.17 , 0.06] | -0.04 | [-0.16 , 0.08] | ||||
| Radboud University | -0.21 | [-0.28 , -0.13] | -0.20 | [-0.27 , -0.12] | ||||
| University of Groningen | 0.61 | [ 0.46 , 0.76] | 0.61 | [ 0.46 , 0.76] | ||||
| Delft University of Technology | 0.37 | [ 0.16 , 0.57] | 0.39 | [ 0.19 , 0.60] | ||||
| Eindhoven University of Technology | 0.50 | [ 0.34 , 0.66] | 0.52 | [ 0.35 , 0.68] | ||||
| Tilburg University | -0.44 | [-0.58 , -0.29] | -0.42 | [-0.56 , -0.27] | ||||
| Maastricht University | -0.02 | [-0.11 , 0.06] | 0.00 | [-0.09 , 0.08] | ||||
| University of Twente | -0.61 | [-0.69 , -0.52] | -0.58 | [-0.67 , -0.50] | ||||
| Utrecht University | -0.18 | [-0.27 , -0.10] | -0.16 | [-0.25 , -0.08] | ||||
| University of Amsterdam | -0.05 | [-0.14 , 0.04] | -0.03 | [-0.12 , 0.05] | ||||
| Vrije Universiteit Amsterdam | -0.05 | [-0.14 , 0.04] | -0.03 | [-0.12 , 0.06] | ||||
| Wageningen University and Research Centre | 0.00 | [-0.09 , 0.08] | 0.01 | [-0.07 , 0.10] | ||||
| Field: ref=Biological and Health Sciences | ||||||||
| Physical and mathematical sciences | -0.20 | [-0.25 , -0.16] | -0.20 | [-0.25 , -0.16] | ||||
| Social and behavioral sciences | 0.19 | [ 0.15 , 0.24] | 0.19 | [ 0.14 , 0.24] | ||||
| Engineering | -0.12 | [-0.18 , -0.06] | -0.12 | [-0.18 , -0.06] | ||||
| Agricultural sciences | 0.05 | [-0.01 , 0.11] | 0.04 | [-0.01 , 0.10] | ||||
| Humanities | 0.29 | [ 0.21 , 0.38] | 0.29 | [ 0.21 , 0.38] | ||||
| Missing field | 0.29 | [ 0.24 , 0.34] | 0.29 | [ 0.24 , 0.34] | ||||
| PhD cohort | -0.03 | [-0.04 , -0.03] | -0.03 | [-0.04 , -0.03] | ||||
| Previous publications | 1.50 | [ 1.43 , 1.57] | 1.50 | [ 1.43 , 1.56] | ||||
| Cohort interactions | ||||||||
| PhD cohort * women | -0.01 | [-0.01 , 0.00] | ||||||
| PhD cohort * missing gender | 0.01 | [ 0.01 , 0.02] | ||||||
| PhD cohort * ethnic minority | 0.00 | [-0.02 , 0.02] | ||||||
| PhD cohort * other ethnicity | 0.00 | [-0.01 , 0.00] | ||||||
| AIC | 65138 | 65451 | 60636 | 60609 | ||||
| N | 15021 | 15021 | 15021 | 15021 |
Robustness checks
R1: 5-year publication window
R1 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender , data = df_ppf5, dist = "lognormal")
R1
R2 <- flexsurvreg(Surv((time -1), time, inactive) ~ ethnicity2 , data = df_ppf5, dist = "lognormal")
R2
R3 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s, data = df_ppf5, dist = "lognormal")
R3
R4 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_ppf5, dist = "lognormal")
R4AMEs Model 3 (no SEs)
dfmaj <- dfmin <- dfmen <- dfwomen <- df_ppf5
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity2 <- "minority" # one dataset all minority
dfmaj$ethnicity2 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df_ppf5$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df_ppf5$gender))
dfmin$ethnicity2 <- factor(dfmin$ethnicity2, levels=levels(df_ppf5$ethnicity2))
dfmaj$ethnicity2 <- factor(dfmaj$ethnicity2, levels=levels(df_ppf5$ethnicity2))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(R3, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(R3, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(R3, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(R3, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
AME_gender
AME_ethnicityAMEs Model 4 (no SEs)
dfmaj <- dfmin <- dfmen <- dfwomen <- df_ppf5
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity2 <- "minority" # one dataset all minority
dfmaj$ethnicity2 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df_ppf5$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df_ppf5$gender))
dfmin$ethnicity2 <- factor(dfmin$ethnicity2, levels=levels(df_ppf5$ethnicity2))
dfmaj$ethnicity2 <- factor(dfmaj$ethnicity2, levels=levels(df_ppf5$ethnicity2))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(R4, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(R4, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(R4, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(R4, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
# cohort interactions
s <- 0.1
# copying our data into separate dataframes
dfwomencp <- dfwomencm <- dfmencp <- dfmencm <- df_ppf5
dfmincp <- dfmincm <- dfmajcp <- dfmajcm <- df_ppf5
# assigning gender to each of the dataframes
dfwomencp$gender <- dfwomencm$gender <- "women"
dfmencp$gender <- dfmencm$gender <- "men"
dfwomencp$gender <- factor(dfwomencp$gender, levels=levels(df_ppf5$gender))
dfwomencm$gender <- factor(dfwomencm$gender, levels=levels(df_ppf5$gender))
dfmencp$gender <- factor(dfmencp$gender, levels=levels(df_ppf5$gender))
dfmencm$gender <- factor(dfmencm$gender, levels=levels(df_ppf5$gender))
# assigning ethnicity to the dataframes
dfmincp$ethnicity2 <- dfmincm$ethnicity2 <- "minority"
dfmajcp$ethnicity2 <- dfmajcm$ethnicity2 <- "majority"
dfmincp$ethnicity2 <- factor(dfmincp$ethnicity2, levels=levels(df_ppf5$ethnicity2))
dfmincm$ethnicity2 <- factor(dfmincm$ethnicity2, levels=levels(df_ppf5$ethnicity2))
dfmajcp$ethnicity2 <- factor(dfmajcp$ethnicity2, levels=levels(df_ppf5$ethnicity2))
dfmajcm$ethnicity2 <- factor(dfmajcm$ethnicity2, levels=levels(df_ppf5$ethnicity2))
# adding/subtracting small changes PhD cohort
dfwomencp$phd_cohort <- dfmencp$phd_cohort <- df_ppf5$phd_cohort + s
dfwomencm$phd_cohort <- dfmencm$phd_cohort <- df_ppf5$phd_cohort - s
dfmajcp$phd_cohort <- dfmincp$phd_cohort <- df_ppf5$phd_cohort + s
dfmajcm$phd_cohort <- dfmincm$phd_cohort <- df_ppf5$phd_cohort - s
# calculating predicted probabilities
pwp <- as.numeric(unlist(predict(R4, type="response", newdata=dfwomencp)))
pwm <- as.numeric(unlist(predict(R4, type="response", newdata=dfwomencm)))
pmp <- as.numeric(unlist(predict(R4, type="response", newdata=dfmencp)))
pmm <- as.numeric(unlist(predict(R4, type="response", newdata=dfmencm)))
pnp <- as.numeric(unlist(predict(R4, type="response", newdata=dfmincp)))
pnm <- as.numeric(unlist(predict(R4, type="response", newdata=dfmincm)))
pjp <- as.numeric(unlist(predict(R4, type="response", newdata=dfmajcp)))
pjm <- as.numeric(unlist(predict(R4, type="response", newdata=dfmajcm)))
# marginal effects
AME_gendercoh <- mean(((pwp - pmp) - (pwm - pmm)) / (2*s))
AME_ethnicitycoh <- mean(((pnp - pjp) - (pnm - pjm)) / (2*s))Model 3 SEs
bootFunc <- function(data, i) {
df <- data[i,] #bootstrap datasets
M <- flexsurv::flexsurvreg(survival::Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s, data = df, dist = "lognormal")
suppressWarnings({
dfmaj <- dfmin <- dfmen <- dfwomen <- df
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity2 <- "minority" # one dataset all minority
dfmaj$ethnicity2 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df$gender))
dfmin$ethnicity2 <- factor(dfmin$ethnicity2, levels=levels(df$ethnicity2))
dfmaj$ethnicity2 <- factor(dfmaj$ethnicity2, levels=levels(df$ethnicity2))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(M, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(M, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(M, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(M, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
})
c(AME_gender, AME_ethnicity)
#save results
}
b3 <- boot(df_ppf5, bootFunc, R = 100, parallel="snow", ncpus=10)
fsave(b3, file = "boot3_R1.rda", location = "./results/stopping/robustness/")original <- b3$t0
bias <- colMeans(b3$t) - b3$t0
se <- apply(b3$t, 2, sd)
boot.df3 <- data.frame(original=original, bias=bias, se=se)
row.names(boot.df3) <- c("AME_gender", "AME_ethnicity")
boot.df3$t <- (boot.df3$original / boot.df3$se)
round(boot.df3, 5)## original bias se t
## AME_gender -4.23738 -1.02307 NA NA
## AME_ethnicity -6.24852 -2.96893 NA NAR2: ethnicity-missing included separately
R5 <- flexsurvreg(Surv((time -1), time, inactive) ~ ethnicity3 , data = df_ppf3, dist = "lognormal")
R5
R6 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity3 + uni + field2 + phd_cohort + npubs_prev_s, data = df_ppf3, dist = "lognormal")
R6
R7 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity3 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_ppf3, dist = "lognormal")
R7AMEs Model 3 (no SEs)
dfmaj <- dfmin <- dfmen <- dfwomen <- df_ppf5
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity3 <- "minority" # one dataset all minority
dfmaj$ethnicity3 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df_ppf5$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df_ppf5$gender))
dfmin$ethnicity3 <- factor(dfmin$ethnicity3, levels=levels(df_ppf5$ethnicity3))
dfmaj$ethnicity3 <- factor(dfmaj$ethnicity3, levels=levels(df_ppf5$ethnicity3))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(R6, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(R6, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(R6, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(R6, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
AME_gender
AME_ethnicityAMEs Model 4 (no SEs)
dfmaj <- dfmin <- dfmen <- dfwomen <- df_ppf5
dfwomen$gender <- "women" # one dataset of all women
dfmen$gender <- "men" # one dataset of all men
dfmin$ethnicity3 <- "minority" # one dataset all minority
dfmaj$ethnicity3 <- "majority" # one dataset all majority
dfwomen$gender <- factor(dfwomen$gender, levels=levels(df_ppf5$gender))
dfmen$gender <- factor(dfmen$gender, levels=levels(df_ppf5$gender))
dfmin$ethnicity3 <- factor(dfmin$ethnicity3, levels=levels(df_ppf5$ethnicity3))
dfmaj$ethnicity3 <- factor(dfmaj$ethnicity3, levels=levels(df_ppf5$ethnicity3))
#calculate the predicted probabilities for gender
pw <- as.numeric(unlist(predict(R7, type="response", newdata=dfwomen)))
pm <- as.numeric(unlist(predict(R7, type="response", newdata=dfmen)))
# average marginal effects
AME_gender <- mean(pw - pm)
#calculate the predicted probabilities for ethnicity
pmn <- as.numeric(unlist(predict(R7, type="response", newdata=dfmin)))
pmj <- as.numeric(unlist(predict(R7, type="response", newdata=dfmaj)))
# average marginal effects
AME_ethnicity <- mean(pmn - pmj)
# cohort interactions
s <- 0.1
# copying our data into separate dataframes
dfwomencp <- dfwomencm <- dfmencp <- dfmencm <- df_ppf5
dfmincp <- dfmincm <- dfmajcp <- dfmajcm <- df_ppf5
# assigning gender to each of the dataframes
dfwomencp$gender <- dfwomencm$gender <- "women"
dfmencp$gender <- dfmencm$gender <- "men"
dfwomencp$gender <- factor(dfwomencp$gender, levels=levels(df_ppf5$gender))
dfwomencm$gender <- factor(dfwomencm$gender, levels=levels(df_ppf5$gender))
dfmencp$gender <- factor(dfmencp$gender, levels=levels(df_ppf5$gender))
dfmencm$gender <- factor(dfmencm$gender, levels=levels(df_ppf5$gender))
# assigning ethnicity to the dataframes
dfmincp$ethnicity3 <- dfmincm$ethnicity3 <- "minority"
dfmajcp$ethnicity3 <- dfmajcm$ethnicity3 <- "majority"
dfmincp$ethnicity3 <- factor(dfmincp$ethnicity3, levels=levels(df_ppf5$ethnicity3))
dfmincm$ethnicity3 <- factor(dfmincm$ethnicity3, levels=levels(df_ppf5$ethnicity3))
dfmajcp$ethnicity3 <- factor(dfmajcp$ethnicity3, levels=levels(df_ppf5$ethnicity3))
dfmajcm$ethnicity3 <- factor(dfmajcm$ethnicity3, levels=levels(df_ppf5$ethnicity3))
# adding/subtracting small changes PhD cohort
dfwomencp$phd_cohort <- dfmencp$phd_cohort <- df_ppf5$phd_cohort + s
dfwomencm$phd_cohort <- dfmencm$phd_cohort <- df_ppf5$phd_cohort - s
dfmajcp$phd_cohort <- dfmincp$phd_cohort <- df_ppf5$phd_cohort + s
dfmajcm$phd_cohort <- dfmincm$phd_cohort <- df_ppf5$phd_cohort - s
# calculating predicted probabilities
pwp <- as.numeric(unlist(predict(R7, type="response", newdata=dfwomencp)))
pwm <- as.numeric(unlist(predict(R7, type="response", newdata=dfwomencm)))
pmp <- as.numeric(unlist(predict(R7, type="response", newdata=dfmencp)))
pmm <- as.numeric(unlist(predict(R7, type="response", newdata=dfmencm)))
pnp <- as.numeric(unlist(predict(R7, type="response", newdata=dfmincp)))
pnm <- as.numeric(unlist(predict(R7, type="response", newdata=dfmincm)))
pjp <- as.numeric(unlist(predict(R7, type="response", newdata=dfmajcp)))
pjm <- as.numeric(unlist(predict(R7, type="response", newdata=dfmajcm)))
# marginal effects
AME_gendercoh <- mean(((pwp - pmp) - (pwm - pmm)) / (2*s))
AME_ethnicitycoh <- mean(((pnp - pjp) - (pnm - pjm)) / (2*s))
AME_gender
AME_gendercoh
AME_ethnicity
AME_ethnicitycohR3: discrete-time event history regression
df_ppf3 %>%
group_by(time) %>%
summarise(event = sum(inactive),
total = n()) %>%
mutate(hazard = event/total) %>%
ggplot(aes(x=time, y = log(-log(1-hazard)))) +
geom_point() +
geom_smooth()
# C-log-log linkR8 <- glm(formula = inactive ~ time + gender,
family = binomial(link="cloglog"),
data = df_ppf3)
R9 <- glm(formula = inactive ~ time + ethnicity2,
family = binomial(link="cloglog"),
data = df_ppf3)
R10 <- glm(formula = inactive ~ time + gender + ethnicity2 + uni + field + phd_cohort + npubs_prev_s,
family = binomial(link="cloglog"),
data = df_ppf3)
R11 <- glm(formula = inactive ~ time + gender + ethnicity2 + uni + field + phd_cohort + npubs_prev_s + gender*phd_cohort + ethnicity2*phd_cohort,
family = binomial(link="cloglog"),
data = df_ppf3)
summary(R8, exp = T)
summary(R9, exp = T)
summary(R10, exp = T)
summary(R11, exp = T)R4: robustness of gender-cohort interaction to choice of cohorts
In cohort 3 (i.e. 1993), Figure 6 shows a divergent gender pattern: women tend to survive much longer than men in this specific cohort. Hence, the observed cohort effect might be due to this exceptional year. We look into this by removing this cohort and rerunning model 4.
# Removing cohort 3
df_nocoh2 <- df_ppf3[df_ppf3$phd_cohort!=2,]
M4_no2 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_nocoh2, dist = "lognormal")
M4_no2
# Removing cohort 0-2
df_nocoh02 <- df_ppf3[df_ppf3$phd_cohort>2,]
M4_no02 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_nocoh02, dist = "lognormal")
M4_no02
# Removing cohort 0-4
df_nocoh04 <- df_ppf3[df_ppf3$phd_cohort>4,]
M4_no04 <- flexsurvreg(Surv((time -1), time, inactive) ~ gender + ethnicity2 + uni + field2 + phd_cohort + npubs_prev_s + phd_cohort*gender + phd_cohort*ethnicity2, data = df_nocoh04, dist = "lognormal")
M4_no04LS0tDQp0aXRsZTogIkFuYWx5c2VzICdjZWFzaW5nIHRvIHB1Ymxpc2gnIg0KZGF0ZTogIkxhc3QgY29tcGlsZWQgb24gYHIgZm9ybWF0KFN5cy50aW1lKCksICclQiwgJVknKWAiDQpvdXRwdXQ6IA0KICBodG1sX2RvY3VtZW50Og0KICAgIGNzczogdHdlYWtzLmNzcw0KICAgIHRvYzogIHRydWUNCiAgICB0b2NfZmxvYXQ6IHRydWUNCiAgICBudW1iZXJfc2VjdGlvbnM6IGZhbHNlDQogICAgY29kZV9mb2xkaW5nOiBzaG93DQogICAgY29kZV9kb3dubG9hZDogeWVzDQoNCi0tLQ0KDQoNClRoaXMgbGFiIGpvdXJuYWwgcmVwbGljYXRlcyB0aGUgYW5hbHlzZXMgZm9yICdjZWFzaW5nIHRvIHB1Ymxpc2gnLiANCg0KICANCg0KLS0tLQ0KDQoNCmBgYHtyLCBlY2hvPUZBTFNFfQ0KDQpybShsaXN0ID0gbHMoKSkNCg0KDQpgYGANCg0KDQoNCiMgQ3VzdG9tIGZ1bmN0aW9ucw0KDQotIGBwYWNrYWdlLmNoZWNrYDogQ2hlY2sgaWYgcGFja2FnZXMgYXJlIGluc3RhbGxlZCAoYW5kIGluc3RhbGwgaWYgbm90KSBpbiBSIChbc291cmNlXShodHRwczovL3ZiYWxpZ2EuZ2l0aHViLmlvL3ZlcmlmeS10aGF0LXItcGFja2FnZXMtYXJlLWluc3RhbGxlZC1hbmQtbG9hZGVkLykpLiAgDQoNCg0KYGBge3IsIHJlc3VsdHM9J2hpZGUnfQ0KDQpmcGFja2FnZS5jaGVjayA8LSBmdW5jdGlvbihwYWNrYWdlcykgew0KICBsYXBwbHkocGFja2FnZXMsIEZVTiA9IGZ1bmN0aW9uKHgpIHsNCiAgICBpZiAoIXJlcXVpcmUoeCwgY2hhcmFjdGVyLm9ubHkgPSBUUlVFKSkgew0KICAgICAgaW5zdGFsbC5wYWNrYWdlcyh4LCBkZXBlbmRlbmNpZXMgPSBUUlVFKQ0KICAgICAgbGlicmFyeSh4LCBjaGFyYWN0ZXIub25seSA9IFRSVUUpDQogICAgfQ0KICB9KQ0KfQ0KDQpmc2F2ZSA8LSBmdW5jdGlvbih4LCBmaWxlLCBsb2NhdGlvbj0iLi9kYXRhL3Byb2Nlc3NlZC8iKSB7DQogIGRhdGVuYW1lIDwtIHN1YnN0cihnc3ViKCJbOi1dIiwgIiIsIFN5cy50aW1lKCkpLCAxLDgpICANCiAgdG90YWxuYW1lIDwtIHBhc3RlKGxvY2F0aW9uLCBkYXRlbmFtZSwgZmlsZSwgc2VwPSIiKQ0KICBzYXZlKHgsIGZpbGUgPSB0b3RhbG5hbWUpICANCn0NCg0KDQpgYGANCg0KDQotLS0gIA0KDQojIFBhY2thZ2VzDQoNCi0gYHN1cnZpdmFsYDogZ2VuZXJhbCBwYWNrYWdlIHRvIGNyZWF0ZSBsaWZlIHRhYmxlcyBhbmQgc3Vydml2YWwgbW9kZWxzLCBuZWVkZWQgZm9yIGZsZXhzdXJ2DQotIGBmbGV4c3VydmA6IGZpdHRpbmcgZmxleGlibGUgc3Vydml2YWwgcmVncmVzc2lvbiBtb2RlbHMNCi0gYGJvb3RgOiBib290c3RyYXBwaW5nIGZvciBTRXMgb2YgQU1Fcw0KLSBgdGlkeXZlcnNlYDogZm9yIGRhdGEgbWFuaXB1bGF0aW9uDQotIGBnZ3Bsb3QyYDogZm9yIGNyZWF0aW5nIGZpZ3VyZXMgNS03DQotIGBnZ3B1YnJgOiBmb3IgY29tYmluaW5nIHR3byBmaWd1cmVzIGluIG9uZSAocGxvdCA1KQ0KLSBga2FibGVFeHRyYWA6IGZvcm1hdHRpbmcgdGFibGVzDQoNCmBgYHtyLCByZXN1bHRzPSdoaWRlJywgd2FybmluZz1GQUxTRX0NCg0KcGFja2FnZXMgPSBjKCJzdXJ2aXZhbCIsICJmbGV4c3VydiIsICJib290IiwgInRpZHl2ZXJzZSIsICJnZ3Bsb3QyIiwgImdncHViciIsICJrYWJsZUV4dHJhIikgDQoNCmZwYWNrYWdlLmNoZWNrKHBhY2thZ2VzKQ0KDQpgYGANCg0KDQotLS0gDQoNCiMgSW5wdXQNCg0KDQoNCldlIHVzZSB0d28gcHJvY2Vzc2VkIGRhdGFzZXRzLg0KDQoqIFtkZl9zdG9wcGluZy5yZGFdKCJodHRwczovL2dpdGh1Yi5jb20vYW1tdWxkZXJzL2FtYXR0ZXJvZnRpbWUvZGF0YS9wcm9jZXNzZWQvZGZfc3RvcHBpbmcucmRhIik6IHBlcnNvbi1wZXJpb2QgZmlsZSBjb250YWluaW5nIHB1YmxpY2F0aW9ucyBhbmQgYWxsIHJlbGV2YW50IHZhcmlhYmxlcyBmb3IgdGhlIHN1cnZpdmFsIG1vZGVscywgdGltZSB3aW5kb3cgZm9yIGluYWN0aXZpdHkgaXMgMyB5ZWFycw0KICAgIC0gRm9yIGNvbnN0cnVjdGlvbiBvZiB0aGlzIGRhdGFzZXQgc2VlIFtEZXBlbmRlbnQgVmFyaWFibGVzIDogU3RhcnRpbmcgYW5kIFN0b3BwaW5nIHRvIFB1Ymxpc2hdKGRhdGFwcmVwYXJhdGlvbi5odG1sKSAgDQogICAgLSBuYW1lIG9mIGRhdGFzZXQ6IGBkZl9wcGYzYCANCg0KKiBbZGZfc3RvcHBpbmc1LnJkYV0oImh0dHBzOi8vZ2l0aHViLmNvbS9hbW11bGRlcnMvYW1hdHRlcm9mdGltZS9kYXRhL3Byb2Nlc3NlZC9kZl9zdG9wcGluZ181LnJkYSIpOiBwZXJzb24tcGVyaW9kIGZpbGUgY29udGFpbmluZyBwdWJsaWNhdGlvbnMgYW5kIGFsbCByZWxldmFudCB2YXJpYWJsZXMgZm9yIHRoZSBzdXJ2aXZhbCBtb2RlbHMsIHRpbWUtd2luZG93IGZvciBpbmFjdGl2aXR5IGlzIHllYXJzDQogICAgLSBGb3IgY29uc3RydWN0aW9uIG9mIHRoaXMgZGF0YXNldCBzZWUgW0RlcGVuZGVudCBWYXJpYWJsZXMgOiBTdGFydGluZyBhbmQgU3RvcHBpbmcgdG8gUHVibGlzaF0oZGF0YXByZXBhcmF0aW9uLmh0bWwpICANCiAgICAtIG5hbWUgb2YgZGF0YXNldDogYGRmX3BwZjVgIA0KDQoNCg0KDQpgYGB7cn0NCg0KbG9hZChmaWxlID0gIi4vZGF0YS9wcm9jZXNzZWQvZGZfc3RvcHBpbmcucmRhIikNCg0KDQpsb2FkKGZpbGUgPSAiZGF0YS9wcm9jZXNzZWQvZGZfc3RvcHBpbmc1LnJkYSIpDQoNCmBgYA0KDQoNCg0KIyBDaGVja2luZyBkaXN0cmlidXRpb25zDQoNCkthcGxhbi1NZWllciBjdXJ2ZQ0KYGBge3J9DQoNCktNMSA8LSBzdXJ2Zml0KFN1cnYodGltZS0xLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBjb25mLmxvd2VyPSd1c3VhbCcsIGRhdGEgPSBkZl9wcGYzKQ0KS00yIDwtIHN1cnZmaXQoU3Vydih0aW1lLTEsIHRpbWUsIGluYWN0aXZlKSB+IGdlbmRlciwgY29uZi5sb3dlcj0ndXN1YWwnLCBkYXRhID0gZGZfcHBmMykNCktNMyA8LSBzdXJ2Zml0KFN1cnYodGltZS0xLCB0aW1lLCBpbmFjdGl2ZSkgfiBldGhuaWNpdHkyLCBjb25mLmxvd2VyPSd1c3VhbCcsIGRhdGEgPSBkZl9wcGYzKQ0KDQoNCnBsb3QoS00xKQ0KcGxvdChLTTIpDQpwbG90KEtNMykNCg0KYGBgDQoNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCiNNQzEgPC0gZmxleHN1cnZyZWcoU3VydigodGltZSAtMSksIHRpbWUsIGluYWN0aXZlKSB+IDEsIGRhdGEgPSBkZl9wcGYzLCBkaXN0ID0gImdlbmdhbW1hIikgIyBjb21tZW50ZWQ6IG5vIGNvbnZlcmdlbmNlDQojTUMyIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJnZW5mIikgICAgICMgY29tbWVudGVkOiBubyBjb252ZXJnZW5jZQ0KTUMzIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJ3ZWlidWxsIikNCk1DNCA8LSBmbGV4c3VydnJlZyhTdXJ2KCh0aW1lIC0xKSwgdGltZSwgaW5hY3RpdmUpIH4gMSwgZGF0YSA9IGRmX3BwZjMsIGRpc3QgPSAiZ2FtbWEiKQ0KTUM1IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJleHAiKQ0KTUM2IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJsbG9naXMiKQ0KTUM3IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiAxLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJnb21wZXJ0eiIpDQpNQzggPC0gZmxleHN1cnZyZWcoU3VydigodGltZSAtMSksIHRpbWUsIGluYWN0aXZlKSB+IDEsIGRhdGEgPSBkZl9wcGYzLCBkaXN0ID0gImxvZ25vcm1hbCIpDQoNCnRlc3RkaXN0IDwtIEFJQyhNQzMsIE1DNCwgTUM1LCBNQzYsIE1DNywgTUM4KQ0KdGVzdGRpc3Rbb3JkZXIodGVzdGRpc3QkQUlDKSxdDQoNCmBgYA0KDQoNCg0KDQoNCiMgTW9kZWwgMTogR2VuZGVyIG9ubHkNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCk0xIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJsb2dub3JtYWwiKQ0KDQpgYGANCg0KYGBge3IsIGVjaG89RkFMU0V9DQoNCmxvYWQoInJlc3VsdHMvc3RvcHBpbmcvMjAyMzA0MDVNMS5yZGEiKQ0KDQpNMSA8LSB4DQoNCnJtKHgpDQoNCmBgYA0KDQpgYGB7cn0NCg0KTTENCg0KYGBgDQoNCg0KUC12YWx1ZXMNCg0KDQpgYGB7ciwgZWNobz1GQUxTRX0NCg0KIyBwLXZhbHVlcw0KDQpNMSRwdmFscyA8LSAyKnBub3JtKC1hYnMoTTEkcmVzWywxXS9NMSRyZXNbLDRdKSkNCg0Kcm91bmQoTTEkcHZhbHMsIDQpDQoNCmBgYA0KDQoNCg0KDQojIE1vZGVsIDI6IEV0aG5pY2l0eSBvbmx5DQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQpNMiA8LSBmbGV4c3VydnJlZyhTdXJ2KCh0aW1lIC0xKSwgdGltZSwgaW5hY3RpdmUpIH4gZXRobmljaXR5MiAsIGRhdGEgPSBkZl9wcGYzLCBkaXN0ID0gImxvZ25vcm1hbCIpDQoNCg0KYGBgDQoNCmBgYHtyLCBlY2hvPUZBTFNFfQ0KDQpsb2FkKCIuL3Jlc3VsdHMvc3RvcHBpbmcvMjAyMzA0MDVNMi5yZGEiKQ0KDQpNMiA8LSB4DQoNCnJtKHgpDQoNCmBgYA0KDQpgYGB7cn0NCg0KTTIgDQoNCmBgYA0KDQoNClAtdmFsdWVzDQoNCg0KYGBge3IsIGVjaG89RkFMU0V9DQoNCiMgQ29tcHV0aW5nIHAtdmFsdWVzDQoNCk0yJHB2YWxzIDwtIDIqcG5vcm0oLWFicyhNMiRyZXNbLDFdL00yJHJlc1ssNF0pKQ0KDQpyb3VuZChNMiRwdmFscywgNCkNCg0KYGBgDQoNCg0KDQoNCg0KIyBNb2RlbCAzOiBHZW5kZXIgYW5kIGV0aG5pY2l0eSwgY29udHJvbHMNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCk0zIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcywgZGF0YSA9IGRmX3BwZjMsIGRpc3QgPSAibG9nbm9ybWFsIikNCg0KYGBgDQoNCmBgYHtyLCBlY2hvPUZBTFNFfQ0KDQpsb2FkKCIuL3Jlc3VsdHMvc3RvcHBpbmcvMjAyMzA0MDVNMy5yZGEiKQ0KDQpNMyA8LSB4DQoNCnJtKHgpDQoNCmBgYA0KDQoNCmBgYHtyfQ0KDQpNMw0KDQpgYGANCg0KDQpQLXZhbHVlcw0KDQoNCmBgYHtyLCBlY2hvPUZBTFNFfQ0KDQojIGNvbXB1dGluZyBwLXZhbHVlcw0KDQpNMyRwdmFscyA8LSAyKnBub3JtKC1hYnMoTTMkcmVzWywxXS9NMyRyZXNbLDRdKSkNCg0Kcm91bmQoTTMkcHZhbHMsIDQpDQoNCmBgYA0KDQoNCg0KIyMgQXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzIChBTUVzKQ0KDQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQpib290RnVuYyA8LSBmdW5jdGlvbihkYXRhLCBpKSB7DQogIGRmIDwtIGRhdGFbaSxdICNib290c3RyYXAgZGF0YXNldHMNCiAgDQogIA0KICBNIDwtIGZsZXhzdXJ2OjpmbGV4c3VydnJlZyhzdXJ2aXZhbDo6U3VydigodGltZSAtMSksIHRpbWUsIGluYWN0aXZlKSB+IGdlbmRlciArIGV0aG5pY2l0eTIgKyB1bmkgKyBmaWVsZDIgKyBwaGRfY29ob3J0ICsgbnB1YnNfcHJldl9zLCBkYXRhID0gZGYsIGRpc3QgPSAibG9nbm9ybWFsIikNCiAgDQogIHN1cHByZXNzV2FybmluZ3Moew0KICANCiAgICBkZm1haiA8LSBkZm1pbiA8LSBkZm1lbiA8LSBkZndvbWVuIDwtIGRmDQogICAgZGZ3b21lbiRnZW5kZXIgPC0gIndvbWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCB3b21lbg0KICAgIGRmbWVuJGdlbmRlciA8LSAibWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCBtZW4NCiAgICBkZm1pbiRldGhuaWNpdHkyIDwtICJtaW5vcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWlub3JpdHkNCiAgICBkZm1haiRldGhuaWNpdHkyIDwtICJtYWpvcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWFqb3JpdHkNCiAgICANCiAgICBkZndvbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbiRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGYkZ2VuZGVyKSkNCiAgICBkZm1lbiRnZW5kZXIgPC0gZmFjdG9yKGRmbWVuJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZiRnZW5kZXIpKQ0KICAgIGRmbWluJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWluJGV0aG5pY2l0eTIsIGxldmVscz1sZXZlbHMoZGYkZXRobmljaXR5MikpDQogICAgZGZtYWokZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtYWokZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCg0KDQogICAgI2NhbGN1bGF0ZSB0aGUgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMgZm9yIGdlbmRlcg0KICAgIHB3IDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmd29tZW4pKSkNCiAgICBwbSA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1lbikpKQ0KICAgIA0KICAgICMgYXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzDQogICAgQU1FX2dlbmRlciA8LSBtZWFuKHB3IC0gcG0pDQogICAgDQogICAgI2NhbGN1bGF0ZSB0aGUgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMgZm9yIGV0aG5pY2l0eQ0KICAgIHBtbiA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbikpKQ0KICAgIHBtaiA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1haikpKQ0KDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZXRobmljaXR5IDwtIG1lYW4ocG1uIC0gcG1qKQ0KICAgIA0KICB9KQ0KICANCiAgYyhBTUVfZ2VuZGVyLCBBTUVfZXRobmljaXR5KQ0KICAgI3NhdmUgcmVzdWx0cw0KfQ0KDQpiMyA8LSBib290KGRmX3BwZjMsIGJvb3RGdW5jLCBSID0gOTk5LCBwYXJhbGxlbD0ic25vdyIsIG5jcHVzPTEwKQ0KDQoNCmZzYXZlKGIzLCBmaWxlID0gImJvb3QzLnJkYSIsIGxvY2F0aW9uID0gIi4vcmVzdWx0cy9zdG9wcGluZy8iKQ0KDQoNCmBgYA0KDQpgYGB7ciwgZWNobz1GQUxTRX0NCg0KbG9hZCgiLi9yZXN1bHRzL3N0b3BwaW5nL2Jvb3QzLnJkYSIpDQpiMyA8LSB4DQpybSh4KQ0KDQpgYGANCg0KDQpgYGB7cn0NCg0Kb3JpZ2luYWwgPC0gYjMkdDANCmJpYXMgPC0gY29sTWVhbnMoYjMkdCkgLSBiMyR0MA0Kc2UgPC0gYXBwbHkoYjMkdCwgMiwgc2QpDQoNCmJvb3QuZGYzIDwtIGRhdGEuZnJhbWUob3JpZ2luYWw9b3JpZ2luYWwsIGJpYXM9Ymlhcywgc2U9c2UpDQpyb3cubmFtZXMoYm9vdC5kZjMpIDwtIGMoIkFNRV9nZW5kZXIiLCAiQU1FX2V0aG5pY2l0eSIpDQoNCmJvb3QuZGYzJHQgPC0gKGJvb3QuZGYzJG9yaWdpbmFsIC8gYm9vdC5kZjMkc2UpDQoNCnJvdW5kKGJvb3QuZGYzLCA1KQ0KDQpgYGANCg0KDQoNCiMgTW9kZWwgNDogR2VuZGVyICsgZXRobmljaXR5ICsgY29ob3J0IHggZ2VuZGVyICsgY29ob3J0IHggZXRobmljaXR5ICsgY29udHJvbHMNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCk00IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcyArIHBoZF9jb2hvcnQqZ2VuZGVyICsgcGhkX2NvaG9ydCpldGhuaWNpdHkyLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJsb2dub3JtYWwiKQ0KDQoNCmBgYA0KDQoNCmBgYHtyLCBlY2hvPUZBTFNFfQ0KDQpsb2FkKCIuL3Jlc3VsdHMvc3RvcHBpbmcvMjAyMzA0MDVNNC5yZGEiKQ0KDQpNNCA8LSB4DQoNCnJtKHgpDQoNCmBgYA0KDQpgYGB7cn0NCg0KTTQNCg0KYGBgDQoNCg0KUC12YWx1ZXMNCg0KDQpgYGB7ciwgZWNobz1GQUxTRX0NCg0KIyBjYWxjdWxhdGluZyBwLXZhbHVlcw0KDQpNNCRwdmFscyA8LSAyKnBub3JtKC1hYnMoTTQkcmVzWywxXS9NNCRyZXNbLDRdKSkNCg0Kcm91bmQoTTQkcHZhbHMsIDQpDQoNCmBgYA0KDQoNCg0KIyMgQXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzIChBTUVzKQ0KDQpgYGB7ciwgZXZhbD1GQUxTRX0NCg0KDQpib290RnVuYyA8LSBmdW5jdGlvbihkYXRhLCBpKSB7DQogIGRmIDwtIGRhdGFbaSxdICNib290c3RyYXAgZGF0YXNldHMNCiAgDQogIA0KICBNIDwtIGZsZXhzdXJ2OjpmbGV4c3VydnJlZyhzdXJ2aXZhbDo6U3VydigodGltZSAtMSksIHRpbWUsIGluYWN0aXZlKSB+IGdlbmRlciArIGV0aG5pY2l0eTIgKyB1bmkgKyBmaWVsZDIgKyBwaGRfY29ob3J0ICsgbnB1YnNfcHJldl9zICsgcGhkX2NvaG9ydCpnZW5kZXIgKyBwaGRfY29ob3J0KmV0aG5pY2l0eTIsIGRhdGEgPSBkZiwgZGlzdCA9ICJsb2dub3JtYWwiKQ0KICANCiAgc3VwcHJlc3NXYXJuaW5ncyh7DQogIA0KICAgIGRmbWFqIDwtIGRmbWluIDwtIGRmbWVuIDwtIGRmd29tZW4gPC0gZGYNCiAgICBkZndvbWVuJGdlbmRlciA8LSAid29tZW4iICMgb25lIGRhdGFzZXQgb2YgYWxsIHdvbWVuDQogICAgZGZtZW4kZ2VuZGVyIDwtICJtZW4iICMgb25lIGRhdGFzZXQgb2YgYWxsIG1lbg0KICAgIGRmbWluJGV0aG5pY2l0eTIgPC0gIm1pbm9yaXR5IiAjIG9uZSBkYXRhc2V0IGFsbCBtaW5vcml0eQ0KICAgIGRmbWFqJGV0aG5pY2l0eTIgPC0gIm1ham9yaXR5IiAjIG9uZSBkYXRhc2V0IGFsbCBtYWpvcml0eQ0KICAgIA0KICAgIGRmd29tZW4kZ2VuZGVyIDwtIGZhY3RvcihkZndvbWVuJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZiRnZW5kZXIpKQ0KICAgIGRmbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZtZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmJGdlbmRlcikpDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtaW4kZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCiAgICBkZm1haiRldGhuaWNpdHkyIDwtIGZhY3RvcihkZm1haiRldGhuaWNpdHkyLCBsZXZlbHM9bGV2ZWxzKGRmJGV0aG5pY2l0eTIpKQ0KDQoNCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZ2VuZGVyDQogICAgcHcgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZ3b21lbikpKQ0KICAgIHBtIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuKSkpDQogICAgDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZ2VuZGVyIDwtIG1lYW4ocHcgLSBwbSkNCiAgICANCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZXRobmljaXR5DQogICAgcG1uIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWluKSkpDQogICAgcG1qIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWFqKSkpDQoNCiAgICAjIGF2ZXJhZ2UgbWFyZ2luYWwgZWZmZWN0cw0KICAgIEFNRV9ldGhuaWNpdHkgPC0gbWVhbihwbW4gLSBwbWopDQogICAgDQogICAgDQogICAgIyBjb2hvcnQgaW50ZXJhY3Rpb25zDQogICAgDQogICAgcyA8LSAwLjENCiAgICAjIGNvcHlpbmcgb3VyIGRhdGEgaW50byBzZXBhcmF0ZSBkYXRhZnJhbWVzDQogICAgZGZ3b21lbmNwIDwtIGRmd29tZW5jbSA8LSBkZm1lbmNwIDwtIGRmbWVuY20gPC0gZGYNCiAgICBkZm1pbmNwIDwtIGRmbWluY20gPC0gZGZtYWpjcCA8LSBkZm1hamNtIDwtIGRmDQogICAgDQogICAgIyBhc3NpZ25pbmcgZ2VuZGVyIHRvIGVhY2ggb2YgdGhlIGRhdGFmcmFtZXMNCiAgICBkZndvbWVuY3AkZ2VuZGVyIDwtIGRmd29tZW5jbSRnZW5kZXIgPC0gIndvbWVuIg0KICAgIGRmbWVuY3AkZ2VuZGVyIDwtIGRmbWVuY20kZ2VuZGVyIDwtICJtZW4iDQogICAgZGZ3b21lbmNwJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbmNwJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZiRnZW5kZXIpKQ0KICAgIGRmd29tZW5jbSRnZW5kZXIgPC0gZmFjdG9yKGRmd29tZW5jbSRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGYkZ2VuZGVyKSkNCiAgICBkZm1lbmNwJGdlbmRlciA8LSBmYWN0b3IoZGZtZW5jcCRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGYkZ2VuZGVyKSkNCiAgICBkZm1lbmNtJGdlbmRlciA8LSBmYWN0b3IoZGZtZW5jbSRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGYkZ2VuZGVyKSkNCiAgICANCiAgICAjIGFzc2lnbmluZyBldGhuaWNpdHkgdG8gdGhlIGRhdGFmcmFtZXMNCiAgICBkZm1pbmNwJGV0aG5pY2l0eTIgPC0gZGZtaW5jbSRldGhuaWNpdHkyIDwtICJtaW5vcml0eSINCiAgICBkZm1hamNwJGV0aG5pY2l0eTIgPC0gZGZtYWpjbSRldGhuaWNpdHkyIDwtICJtYWpvcml0eSINCiAgICBkZm1pbmNwJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWluY3AkZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCiAgICBkZm1pbmNtJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWluY20kZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCiAgICBkZm1hamNwJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqY3AkZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCiAgICBkZm1hamNtJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqY20kZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZiRldGhuaWNpdHkyKSkNCiAgICANCiAgICANCiAgICAjIGFkZGluZy9zdWJ0cmFjdGluZyBzbWFsbCBjaGFuZ2VzIFBoRCBjb2hvcnQNCiAgICBkZndvbWVuY3AkcGhkX2NvaG9ydCA8LSBkZm1lbmNwJHBoZF9jb2hvcnQgPC0gZGYkcGhkX2NvaG9ydCArIHMNCiAgICBkZndvbWVuY20kcGhkX2NvaG9ydCA8LSBkZm1lbmNtJHBoZF9jb2hvcnQgPC0gZGYkcGhkX2NvaG9ydCAtIHMNCiAgICANCiAgICBkZm1hamNwJHBoZF9jb2hvcnQgPC0gZGZtaW5jcCRwaGRfY29ob3J0IDwtIGRmJHBoZF9jb2hvcnQgKyBzDQogICAgZGZtYWpjbSRwaGRfY29ob3J0IDwtIGRmbWluY20kcGhkX2NvaG9ydCA8LSBkZiRwaGRfY29ob3J0IC0gcw0KICAgIA0KICAgICMgY2FsY3VsYXRpbmcgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMNCiAgICBwd3AgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZ3b21lbmNwKSkpDQogICAgcHdtIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmd29tZW5jbSkpKQ0KICAgIHBtcCA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1lbmNwKSkpDQogICAgcG1tIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuY20pKSkNCiAgICANCiAgICBwbnAgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtaW5jcCkpKQ0KICAgIHBubSA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbmNtKSkpDQogICAgcGpwIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoTSwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWFqY3ApKSkNCiAgICBwam0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtYWpjbSkpKQ0KICAgIA0KICAgIA0KIyBtYXJnaW5hbCBlZmZlY3RzDQpBTUVfZ2VuZGVyY29oIDwtIG1lYW4oKChwd3AgLSBwbXApIC0gKHB3bSAtIHBtbSkpIC8gKDIqcykpDQoNCkFNRV9ldGhuaWNpdHljb2ggPC0gbWVhbigoKHBucCAtIHBqcCkgLSAocG5tIC0gcGptKSkgLyAoMipzKSkNCg0KICB9KQ0KICANCiAgYyhBTUVfZ2VuZGVyLCBBTUVfZXRobmljaXR5LCBBTUVfZ2VuZGVyY29oLCBBTUVfZXRobmljaXR5Y29oKQ0KICAgI3NhdmUgcmVzdWx0cw0KfQ0KDQpiNCA8LSBib290KGRmX3BwZjMsIGJvb3RGdW5jLCBSID0gOTk5LCBwYXJhbGxlbD0ic25vdyIsIG5jcHVzPTEwKQ0KDQoNCmZzYXZlKGI0LCBmaWxlID0gImJvb3Q0LnJkYSIsIGxvY2F0aW9uID0gIi4vcmVzdWx0cy9zdG9wcGluZy8iKQ0KDQoNCmBgYA0KDQpgYGB7ciwgZWNobz1GQUxTRX0NCg0KbG9hZChmaWxlPSAiLi9yZXN1bHRzL3N0b3BwaW5nLzIwMjMwNDA4Ym9vdDQucmRhIikNCg0KYjQgPC0geA0KDQpybSh4KQ0KDQpgYGANCg0KDQpgYGB7cn0NCg0Kb3JpZ2luYWwgPC0gYjQkdDANCmJpYXMgPC0gY29sTWVhbnMoYjQkdCkgLSBiNCR0MA0Kc2UgPC0gYXBwbHkoYjQkdCwgMiwgc2QpDQoNCmJvb3QuZGY0IDwtIGRhdGEuZnJhbWUob3JpZ2luYWw9b3JpZ2luYWwsIGJpYXM9Ymlhcywgc2U9c2UpDQpyb3cubmFtZXMoYm9vdC5kZjQpIDwtIGMoIkFNRV9nZW5kZXIiLCAiQU1FX2V0aG5pY2l0eSIsICJBTUVfZ2VuZGVyLWNvaCIsICJBTUVfZXRobmljaXR5LWNvaCIpDQoNCmJvb3QuZGY0JHQgPC0gKGJvb3QuZGY0JG9yaWdpbmFsIC8gYm9vdC5kZjQkc2UpDQoNCnJvdW5kKGJvb3QuZGY0LCA1KQ0KDQpgYGANCg0KDQoNCiMgVGFibGUgNA0KDQoNCmBgYHtyfQ0KDQpjb2x1bW5zIDwtIGMocmVwKGMoIkIiLCAiQy5JLiIpLCA0KSkNCnJvd3MgPC0gYygiPHN0cm9uZz5TaGFwZTwvc3Ryb25nPjwvcD4iLCAiPHN0cm9uZz5TY2FsZTwvc3Ryb25nPjwvcD4iLCAiR2VuZGVyOiByZWY9bWVuIiAsIldvbWVuIiwgIk1pc3NpbmcgZ2VuZGVyIiwgIkV0aG5pY2l0eTogcmVmPW1ham9yaXR5IiwiTWlub3JpdHkiLCAiT3RoZXIiLCAiVW5pdmVyc2l0eTogcmVmPUVyYXNtdXMgVW5pdmVyc2l0eSIsICJMZWlkZW4gVW5pdmVyc2l0eSIsICJSYWRib3VkIFVuaXZlcnNpdHkiLCAiVW5pdmVyc2l0eSBvZiBHcm9uaW5nZW4iLCAiRGVsZnQgVW5pdmVyc2l0eSBvZiBUZWNobm9sb2d5IiwgIkVpbmRob3ZlbiBVbml2ZXJzaXR5IG9mIFRlY2hub2xvZ3kiLCAiVGlsYnVyZyBVbml2ZXJzaXR5IiwgIk1hYXN0cmljaHQgVW5pdmVyc2l0eSIsICJVbml2ZXJzaXR5IG9mIFR3ZW50ZSIsICJVdHJlY2h0IFVuaXZlcnNpdHkiLCAiVW5pdmVyc2l0eSBvZiBBbXN0ZXJkYW0iLCAiVnJpamUgVW5pdmVyc2l0ZWl0IEFtc3RlcmRhbSIsICJXYWdlbmluZ2VuIFVuaXZlcnNpdHkgYW5kIFJlc2VhcmNoIENlbnRyZSIsICJGaWVsZDogcmVmPUJpb2xvZ2ljYWwgYW5kIEhlYWx0aCBTY2llbmNlcyIsICJQaHlzaWNhbCBhbmQgbWF0aGVtYXRpY2FsIHNjaWVuY2VzIiwgIlNvY2lhbCBhbmQgYmVoYXZpb3JhbCBzY2llbmNlcyIsICJFbmdpbmVlcmluZyIsICJBZ3JpY3VsdHVyYWwgc2NpZW5jZXMiLCAiSHVtYW5pdGllcyIsICJNaXNzaW5nIGZpZWxkIiwgIjxzdHJvbmc+UGhEIGNvaG9ydDwvc3Ryb25nPjwvcD4iLCAiPHN0cm9uZz5QcmV2aW91cyBwdWJsaWNhdGlvbnM8L3N0cm9uZz48L3A+IiwgIkNvaG9ydCBpbnRlcmFjdGlvbnMiLCJQaEQgY29ob3J0ICogd29tZW4iLCAiUGhEIGNvaG9ydCAqIG1pc3NpbmcgZ2VuZGVyIiwgIlBoRCBjb2hvcnQgKiBldGhuaWMgbWlub3JpdHkiLCAiUGhEIGNvaG9ydCAqIG90aGVyIGV0aG5pY2l0eSIsICI8c3Ryb25nPkFJQyIsICI8c3Ryb25nPk4iKQ0KDQoNCnQ0IDwtIGRhdGEuZnJhbWUobWF0cml4KG5yb3c9bGVuZ3RoKHJvd3MpLCBuY29sPWxlbmd0aChjb2x1bW5zKSkpDQpjb2xuYW1lcyh0NCkgPC0gY29sdW1ucw0Kcm93bmFtZXModDQpIDwtIHJvd3MNCg0KDQoNCiMgTW9kZWwgMQ0KdDRbYygxLDIsNCw1KSwxXSA8LSBmb3JtYXQocm91bmQoYXMubnVtZXJpYyhNMSRjb2VmKSwgMiksIG5zbWFsbD0yKSAjIGVzdGltYXRlcw0KdDRbYygxLDIpLDJdIDwtIHBhc3RlMCgiWyIsIGZvcm1hdChyb3VuZChsb2coTTEkcmVzW2MoMSwyKSwyXSksIDIpLCBuc21hbGw9MiksICIgLCAiLCBmb3JtYXQocm91bmQobG9nKE0xJHJlc1tjKDEsMiksM10pLCAyKSwgbnNtYWxsPTIpLCAiXSIpICMgQ0kgZm9yIHNoYXBlIGFuZCBzY2FsZQ0KdDRbYyg0LDUpLDJdIDwtIHBhc3RlMCgiWyIsIGZvcm1hdChyb3VuZChNMSRyZXNbYygzLDQpLDJdLCAyKSwgbnNtYWxsPTIpLCAiICwgIiwgZm9ybWF0KHJvdW5kKE0xJHJlc1tjKDMsNCksM10sIDMpLCBuc21hbGw9MiksICJdIikgIyBDSSBmb3Igb3RoZXIgY292YXJpYXRlcw0KDQoNCiMgTW9kZWwgMg0KdDRbYygxLDIsNyw4KSwzXSA8LSBmb3JtYXQocm91bmQoTTIkY29lZiwgMiksIG5zbWFsbD0yKQ0KdDRbYygxLDIpLDRdIDwtIHBhc3RlMCgiWyIsIGZvcm1hdChyb3VuZChsb2coTTIkcmVzW2MoMSwyKSwyXSksIDIpLCBuc21hbGw9MiksICIgLCAiLCBmb3JtYXQocm91bmQobG9nKE0yJHJlc1tjKDEsMiksM10pLCAyKSwgbnNtYWxsPTIpLCAiXSIpDQp0NFtjKDcsOCksNF0gPC0gcGFzdGUwKCJbIiwgZm9ybWF0KHJvdW5kKE0yJHJlc1tjKDMsNCksMl0sIDIpLCBuc21hbGw9MiksICIgLCAiLCBmb3JtYXQocm91bmQoTTIkcmVzW2MoMyw0KSwzXSwgMiksIG5zbWFsbD0yKSwgIl0iKQ0KDQojIE1vZGVsIDMNCnQ0W2MoMSwyLDQsNSw3LDgsMTA6MjEsMjM6MzApLDVdIDwtIGZvcm1hdChyb3VuZChNMyRjb2VmLCAyKSwgbnNtYWxsPTIpDQojdDRbYygxLDIpLCA2XSA8LSBwYXN0ZSgiWyIsIHJvdW5kKGxvZyhNMyRyZXNbYygxLDIpLDJdKSwgMyksICIsIiwgcm91bmQobG9nKE0zJHJlc1tjKDEsMiksM10pLCAzKSwgIl0iKQ0KdDRbYygxLDIsNCw1LDcsOCwxMDoyMSwyMzozMCksIDZdIDwtIHBhc3RlMCgiWyIsIGZvcm1hdChyb3VuZChNMyRyZXNbYygxLDIsMzoyNiksMl0sIDIpLCBuc21hbGw9MiksICIgLCAiLCBmb3JtYXQocm91bmQoTTMkcmVzW2MoMSwyLDM6MjYpLDNdLCAyKSwgbnNtYWxsPTIpLCAiXSIpDQoNCiMgTW9kZWwgNCANCnQ0W2MoMSwyLDQsNSw3LDgsMTA6MjEsMjM6MzAsMzI6MzUpLDddIDwtIGZvcm1hdChyb3VuZChNNCRjb2VmLCAyKSwgbnNtYWxsPTIpDQojdDRbYygxLDIpLCA4XSA8LSBwYXN0ZSgiWyIsIHJvdW5kKGxvZyhNNCRyZXNbYygxOjIpLDJdKSwgMyksICIsIiwgcm91bmQobG9nKE00JHJlc1tjKDE6MiksM10pLCAzKSwgIl0iKQ0KdDRbYygxLDIsNCw1LDcsOCwxMDoyMSwyMzozMCwzMjozNSksIDhdIDwtIHBhc3RlMCgiWyIsIGZvcm1hdChyb3VuZChNNCRyZXNbYygxLDIsMzozMCksMl0sIDIpLCBuc21hbGw9MiksICIgLCAiLCBmb3JtYXQocm91bmQoTTQkcmVzW2MoMSwyLDM6MzApLDNdLCAyKSwgbnNtYWxsPTIpLCAiXSIpDQoNCg0KIyBBSUMNCnQ0WzM2LDFdIDwtIHJvdW5kKE0xJEFJQywgMCkNCnQ0WzM2LDNdIDwtIHJvdW5kKE0yJEFJQywgMCkNCnQ0WzM2LDVdIDwtIHJvdW5kKE0zJEFJQywgMCkNCnQ0WzM2LDddIDwtIHJvdW5kKE00JEFJQywgMCkNCg0KIyBzYW1wbGUgc2l6ZQ0KdDRbMzcsIGMoMSwzLDUsNyldIDwtIHJlcChucm93KGRmX3BwZjNbIWR1cGxpY2F0ZWQoZGZfcHBmMyRpZCksXSksIDQpDQoNCg0KdDRbaXMubmEodDQpXSA8LSAiIg0KDQoNCmBgYA0KDQoNCmBgYHtyfQ0KDQp0NCAlPiUNCiAga2FibGUoZm9ybWF0ID0gJ2h0bWwnLCBjYXB0aW9uID0gJzxiPlRhYmxlIDQuPC9iPiBMb2ctbm9ybWFsIHJlZ3Jlc3Npb24gYW5hbHlzZXMgb24gc3RvcHBpbmcgdG8gcHVibGlzaCcsIGVzY2FwZT1GQUxTRSkgJT4lDQogIGFkZF9oZWFkZXJfYWJvdmUoLixjKCcgJz0xLCAnTW9kZWwgMSc9MiwgJ01vZGVsIDInPTIsICdNb2RlbCAzJz0yLCAnTW9kZWwgNCc9MiksIGVzY2FwZSA9IEZBTFNFLCBib2xkID0gVFJVRSkgJT4lDQogIHJvd19zcGVjKHJvdz1jKDMsNiw5LDIyLDMxKSwgYm9sZD1UKSAlPiUNCiAga2FibGVfY2xhc3NpYyhmdWxsX3dpZHRoID0gRiwgaHRtbF9mb250ID0gIkNhbWJyaWEiKSAlPiUNCiAga2FibGVfc3R5bGluZyhmb250X3NpemUgPSAxMSkgLT4gdGFibGU0DQoNCnRhYmxlNA0KDQpgYGANCg0KDQoNCg0KIyBSb2J1c3RuZXNzIGNoZWNrcw0KDQojIyBSMTogNS15ZWFyIHB1YmxpY2F0aW9uIHdpbmRvdw0KDQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQpSMSA8LSBmbGV4c3VydnJlZyhTdXJ2KCh0aW1lIC0xKSwgdGltZSwgaW5hY3RpdmUpIH4gZ2VuZGVyICwgZGF0YSA9IGRmX3BwZjUsIGRpc3QgPSAibG9nbm9ybWFsIikNClIxDQoNClIyIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBldGhuaWNpdHkyICwgZGF0YSA9IGRmX3BwZjUsIGRpc3QgPSAibG9nbm9ybWFsIikNClIyDQoNClIzIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcywgZGF0YSA9IGRmX3BwZjUsIGRpc3QgPSAibG9nbm9ybWFsIikNClIzDQoNClI0IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcyArIHBoZF9jb2hvcnQqZ2VuZGVyICsgcGhkX2NvaG9ydCpldGhuaWNpdHkyLCBkYXRhID0gZGZfcHBmNSwgZGlzdCA9ICJsb2dub3JtYWwiKQ0KUjQNCg0KYGBgDQoNCg0KDQoNCkFNRXMgTW9kZWwgMyAobm8gU0VzKQ0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCmRmbWFqIDwtIGRmbWluIDwtIGRmbWVuIDwtIGRmd29tZW4gPC0gZGZfcHBmNQ0KICAgIGRmd29tZW4kZ2VuZGVyIDwtICJ3b21lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgd29tZW4NCiAgICBkZm1lbiRnZW5kZXIgPC0gIm1lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgbWVuDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSAibWlub3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1pbm9yaXR5DQogICAgZGZtYWokZXRobmljaXR5MiA8LSAibWFqb3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1ham9yaXR5DQogICAgDQogICAgZGZ3b21lbiRnZW5kZXIgPC0gZmFjdG9yKGRmd29tZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1lbiRnZW5kZXIgPC0gZmFjdG9yKGRmbWVuJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGdlbmRlcikpDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtaW4kZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTIpKQ0KICAgIGRmbWFqJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqJGV0aG5pY2l0eTIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRldGhuaWNpdHkyKSkNCg0KDQogICAgI2NhbGN1bGF0ZSB0aGUgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMgZm9yIGdlbmRlcg0KICAgIHB3IDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjMsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZndvbWVuKSkpDQogICAgcG0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSMywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuKSkpDQogICAgDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZ2VuZGVyIDwtIG1lYW4ocHcgLSBwbSkNCiAgICANCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZXRobmljaXR5DQogICAgcG1uIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjMsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbikpKQ0KICAgIHBtaiA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFIzLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtYWopKSkNCg0KICAgICMgYXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzDQogICAgQU1FX2V0aG5pY2l0eSA8LSBtZWFuKHBtbiAtIHBtaikNCiAgICANCiAgICANCkFNRV9nZW5kZXINCkFNRV9ldGhuaWNpdHkNCg0KYGBgDQoNCkFNRXMgTW9kZWwgNCAobm8gU0VzKQ0KDQpgYGB7ciwgZXZhbD1GQUxTRX0NCg0KICAgIGRmbWFqIDwtIGRmbWluIDwtIGRmbWVuIDwtIGRmd29tZW4gPC0gZGZfcHBmNQ0KICAgIGRmd29tZW4kZ2VuZGVyIDwtICJ3b21lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgd29tZW4NCiAgICBkZm1lbiRnZW5kZXIgPC0gIm1lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgbWVuDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSAibWlub3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1pbm9yaXR5DQogICAgZGZtYWokZXRobmljaXR5MiA8LSAibWFqb3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1ham9yaXR5DQogICAgDQogICAgZGZ3b21lbiRnZW5kZXIgPC0gZmFjdG9yKGRmd29tZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1lbiRnZW5kZXIgPC0gZmFjdG9yKGRmbWVuJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGdlbmRlcikpDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtaW4kZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTIpKQ0KICAgIGRmbWFqJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqJGV0aG5pY2l0eTIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRldGhuaWNpdHkyKSkNCg0KDQogICAgI2NhbGN1bGF0ZSB0aGUgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMgZm9yIGdlbmRlcg0KICAgIHB3IDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZndvbWVuKSkpDQogICAgcG0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNCwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuKSkpDQogICAgDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZ2VuZGVyIDwtIG1lYW4ocHcgLSBwbSkNCiAgICANCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZXRobmljaXR5DQogICAgcG1uIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbikpKQ0KICAgIHBtaiA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFI0LCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtYWopKSkNCg0KICAgICMgYXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzDQogICAgQU1FX2V0aG5pY2l0eSA8LSBtZWFuKHBtbiAtIHBtaikNCiAgICANCiAgICANCiAgICAjIGNvaG9ydCBpbnRlcmFjdGlvbnMNCiAgICANCiAgICBzIDwtIDAuMQ0KICAgICMgY29weWluZyBvdXIgZGF0YSBpbnRvIHNlcGFyYXRlIGRhdGFmcmFtZXMNCiAgICBkZndvbWVuY3AgPC0gZGZ3b21lbmNtIDwtIGRmbWVuY3AgPC0gZGZtZW5jbSA8LSBkZl9wcGY1DQogICAgZGZtaW5jcCA8LSBkZm1pbmNtIDwtIGRmbWFqY3AgPC0gZGZtYWpjbSA8LSBkZl9wcGY1DQogICAgDQogICAgIyBhc3NpZ25pbmcgZ2VuZGVyIHRvIGVhY2ggb2YgdGhlIGRhdGFmcmFtZXMNCiAgICBkZndvbWVuY3AkZ2VuZGVyIDwtIGRmd29tZW5jbSRnZW5kZXIgPC0gIndvbWVuIg0KICAgIGRmbWVuY3AkZ2VuZGVyIDwtIGRmbWVuY20kZ2VuZGVyIDwtICJtZW4iDQogICAgZGZ3b21lbmNwJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbmNwJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGdlbmRlcikpDQogICAgZGZ3b21lbmNtJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbmNtJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGdlbmRlcikpDQogICAgZGZtZW5jcCRnZW5kZXIgPC0gZmFjdG9yKGRmbWVuY3AkZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1lbmNtJGdlbmRlciA8LSBmYWN0b3IoZGZtZW5jbSRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRnZW5kZXIpKQ0KICAgIA0KICAgICMgYXNzaWduaW5nIGV0aG5pY2l0eSB0byB0aGUgZGF0YWZyYW1lcw0KICAgIGRmbWluY3AkZXRobmljaXR5MiA8LSBkZm1pbmNtJGV0aG5pY2l0eTIgPC0gIm1pbm9yaXR5Ig0KICAgIGRmbWFqY3AkZXRobmljaXR5MiA8LSBkZm1hamNtJGV0aG5pY2l0eTIgPC0gIm1ham9yaXR5Ig0KICAgIGRmbWluY3AkZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtaW5jcCRldGhuaWNpdHkyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZXRobmljaXR5MikpDQogICAgZGZtaW5jbSRldGhuaWNpdHkyIDwtIGZhY3RvcihkZm1pbmNtJGV0aG5pY2l0eTIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRldGhuaWNpdHkyKSkNCiAgICBkZm1hamNwJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqY3AkZXRobmljaXR5MiwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTIpKQ0KICAgIGRmbWFqY20kZXRobmljaXR5MiA8LSBmYWN0b3IoZGZtYWpjbSRldGhuaWNpdHkyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZXRobmljaXR5MikpDQogICAgDQogICAgDQogICAgIyBhZGRpbmcvc3VidHJhY3Rpbmcgc21hbGwgY2hhbmdlcyBQaEQgY29ob3J0DQogICAgZGZ3b21lbmNwJHBoZF9jb2hvcnQgPC0gZGZtZW5jcCRwaGRfY29ob3J0IDwtIGRmX3BwZjUkcGhkX2NvaG9ydCArIHMNCiAgICBkZndvbWVuY20kcGhkX2NvaG9ydCA8LSBkZm1lbmNtJHBoZF9jb2hvcnQgPC0gZGZfcHBmNSRwaGRfY29ob3J0IC0gcw0KICAgIA0KICAgIGRmbWFqY3AkcGhkX2NvaG9ydCA8LSBkZm1pbmNwJHBoZF9jb2hvcnQgPC0gZGZfcHBmNSRwaGRfY29ob3J0ICsgcw0KICAgIGRmbWFqY20kcGhkX2NvaG9ydCA8LSBkZm1pbmNtJHBoZF9jb2hvcnQgPC0gZGZfcHBmNSRwaGRfY29ob3J0IC0gcw0KICAgIA0KICAgICMgY2FsY3VsYXRpbmcgcHJlZGljdGVkIHByb2JhYmlsaXRpZXMNCiAgICBwd3AgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNCwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmd29tZW5jcCkpKQ0KICAgIHB3bSA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFI0LCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZ3b21lbmNtKSkpDQogICAgcG1wIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1lbmNwKSkpDQogICAgcG1tIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1lbmNtKSkpDQogICAgDQogICAgcG5wIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbmNwKSkpDQogICAgcG5tIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1pbmNtKSkpDQogICAgcGpwIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1hamNwKSkpDQogICAgcGptIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjQsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1hamNtKSkpDQogICAgDQogICAgDQojIG1hcmdpbmFsIGVmZmVjdHMNCkFNRV9nZW5kZXJjb2ggPC0gbWVhbigoKHB3cCAtIHBtcCkgLSAocHdtIC0gcG1tKSkgLyAoMipzKSkNCg0KQU1FX2V0aG5pY2l0eWNvaCA8LSBtZWFuKCgocG5wIC0gcGpwKSAtIChwbm0gLSBwam0pKSAvICgyKnMpKQ0KDQpgYGANCg0KDQoNCg0KTW9kZWwgMyBTRXMNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCg0KYm9vdEZ1bmMgPC0gZnVuY3Rpb24oZGF0YSwgaSkgew0KICBkZiA8LSBkYXRhW2ksXSAjYm9vdHN0cmFwIGRhdGFzZXRzDQogIA0KICANCiAgTSA8LSBmbGV4c3Vydjo6ZmxleHN1cnZyZWcoc3Vydml2YWw6OlN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcywgZGF0YSA9IGRmLCBkaXN0ID0gImxvZ25vcm1hbCIpDQogIA0KICBzdXBwcmVzc1dhcm5pbmdzKHsNCiAgDQogICAgZGZtYWogPC0gZGZtaW4gPC0gZGZtZW4gPC0gZGZ3b21lbiA8LSBkZg0KICAgIGRmd29tZW4kZ2VuZGVyIDwtICJ3b21lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgd29tZW4NCiAgICBkZm1lbiRnZW5kZXIgPC0gIm1lbiIgIyBvbmUgZGF0YXNldCBvZiBhbGwgbWVuDQogICAgZGZtaW4kZXRobmljaXR5MiA8LSAibWlub3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1pbm9yaXR5DQogICAgZGZtYWokZXRobmljaXR5MiA8LSAibWFqb3JpdHkiICMgb25lIGRhdGFzZXQgYWxsIG1ham9yaXR5DQogICAgDQogICAgZGZ3b21lbiRnZW5kZXIgPC0gZmFjdG9yKGRmd29tZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmJGdlbmRlcikpDQogICAgZGZtZW4kZ2VuZGVyIDwtIGZhY3RvcihkZm1lbiRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGYkZ2VuZGVyKSkNCiAgICBkZm1pbiRldGhuaWNpdHkyIDwtIGZhY3RvcihkZm1pbiRldGhuaWNpdHkyLCBsZXZlbHM9bGV2ZWxzKGRmJGV0aG5pY2l0eTIpKQ0KICAgIGRmbWFqJGV0aG5pY2l0eTIgPC0gZmFjdG9yKGRmbWFqJGV0aG5pY2l0eTIsIGxldmVscz1sZXZlbHMoZGYkZXRobmljaXR5MikpDQoNCg0KICAgICNjYWxjdWxhdGUgdGhlIHByZWRpY3RlZCBwcm9iYWJpbGl0aWVzIGZvciBnZW5kZXINCiAgICBwdyA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KE0sIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZndvbWVuKSkpDQogICAgcG0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtZW4pKSkNCiAgICANCiAgICAjIGF2ZXJhZ2UgbWFyZ2luYWwgZWZmZWN0cw0KICAgIEFNRV9nZW5kZXIgPC0gbWVhbihwdyAtIHBtKQ0KICAgIA0KICAgICNjYWxjdWxhdGUgdGhlIHByZWRpY3RlZCBwcm9iYWJpbGl0aWVzIGZvciBldGhuaWNpdHkNCiAgICBwbW4gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtaW4pKSkNCiAgICBwbWogPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChNLCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtYWopKSkNCg0KICAgICMgYXZlcmFnZSBtYXJnaW5hbCBlZmZlY3RzDQogICAgQU1FX2V0aG5pY2l0eSA8LSBtZWFuKHBtbiAtIHBtaikNCiAgICANCiAgfSkNCiAgDQogIGMoQU1FX2dlbmRlciwgQU1FX2V0aG5pY2l0eSkNCiAgICNzYXZlIHJlc3VsdHMNCn0NCg0KYjMgPC0gYm9vdChkZl9wcGY1LCBib290RnVuYywgUiA9IDEwMCwgcGFyYWxsZWw9InNub3ciLCBuY3B1cz0xMCkNCg0KDQpmc2F2ZShiMywgZmlsZSA9ICJib290M19SMS5yZGEiLCBsb2NhdGlvbiA9ICIuL3Jlc3VsdHMvc3RvcHBpbmcvcm9idXN0bmVzcy8iKQ0KDQoNCmBgYA0KDQpgYGB7ciwgZWNobz1GQUxTRX0NCg0KbG9hZCgiLi9yZXN1bHRzL3N0b3BwaW5nL3JvYnVzdG5lc3MvMjAyNDAxMTVib290M19SMS5yZGEiKQ0KYjMgPC0geA0Kcm0oeCkNCg0KYGBgDQoNCg0KYGBge3J9DQoNCm9yaWdpbmFsIDwtIGIzJHQwDQpiaWFzIDwtIGNvbE1lYW5zKGIzJHQpIC0gYjMkdDANCnNlIDwtIGFwcGx5KGIzJHQsIDIsIHNkKQ0KDQpib290LmRmMyA8LSBkYXRhLmZyYW1lKG9yaWdpbmFsPW9yaWdpbmFsLCBiaWFzPWJpYXMsIHNlPXNlKQ0Kcm93Lm5hbWVzKGJvb3QuZGYzKSA8LSBjKCJBTUVfZ2VuZGVyIiwgIkFNRV9ldGhuaWNpdHkiKQ0KDQpib290LmRmMyR0IDwtIChib290LmRmMyRvcmlnaW5hbCAvIGJvb3QuZGYzJHNlKQ0KDQpyb3VuZChib290LmRmMywgNSkNCg0KYGBgDQoNCg0KDQoNCg0KDQoNCiMjIFIyOiBldGhuaWNpdHktbWlzc2luZyBpbmNsdWRlZCBzZXBhcmF0ZWx5DQoNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNClI1IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBldGhuaWNpdHkzICwgZGF0YSA9IGRmX3BwZjMsIGRpc3QgPSAibG9nbm9ybWFsIikNClI1DQoNClI2IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkzICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcywgZGF0YSA9IGRmX3BwZjMsIGRpc3QgPSAibG9nbm9ybWFsIikNClI2DQoNClI3IDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkzICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcyArIHBoZF9jb2hvcnQqZ2VuZGVyICsgcGhkX2NvaG9ydCpldGhuaWNpdHkyLCBkYXRhID0gZGZfcHBmMywgZGlzdCA9ICJsb2dub3JtYWwiKQ0KUjcNCg0KDQpgYGANCg0KDQoNCg0KQU1FcyBNb2RlbCAzIChubyBTRXMpDQpgYGB7ciwgZXZhbD1GQUxTRX0NCg0KZGZtYWogPC0gZGZtaW4gPC0gZGZtZW4gPC0gZGZ3b21lbiA8LSBkZl9wcGY1DQogICAgZGZ3b21lbiRnZW5kZXIgPC0gIndvbWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCB3b21lbg0KICAgIGRmbWVuJGdlbmRlciA8LSAibWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCBtZW4NCiAgICBkZm1pbiRldGhuaWNpdHkzIDwtICJtaW5vcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWlub3JpdHkNCiAgICBkZm1haiRldGhuaWNpdHkzIDwtICJtYWpvcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWFqb3JpdHkNCiAgICANCiAgICBkZndvbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbiRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRnZW5kZXIpKQ0KICAgIGRmbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZtZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1pbiRldGhuaWNpdHkzIDwtIGZhY3RvcihkZm1pbiRldGhuaWNpdHkzLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZXRobmljaXR5MykpDQogICAgZGZtYWokZXRobmljaXR5MyA8LSBmYWN0b3IoZGZtYWokZXRobmljaXR5MywgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTMpKQ0KDQoNCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZ2VuZGVyDQogICAgcHcgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNiwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmd29tZW4pKSkNCiAgICBwbSA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFI2LCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtZW4pKSkNCiAgICANCiAgICAjIGF2ZXJhZ2UgbWFyZ2luYWwgZWZmZWN0cw0KICAgIEFNRV9nZW5kZXIgPC0gbWVhbihwdyAtIHBtKQ0KICAgIA0KICAgICNjYWxjdWxhdGUgdGhlIHByZWRpY3RlZCBwcm9iYWJpbGl0aWVzIGZvciBldGhuaWNpdHkNCiAgICBwbW4gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNiwgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWluKSkpDQogICAgcG1qIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjYsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1haikpKQ0KDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZXRobmljaXR5IDwtIG1lYW4ocG1uIC0gcG1qKQ0KICAgIA0KICAgIA0KQU1FX2dlbmRlcg0KQU1FX2V0aG5pY2l0eQ0KDQpgYGANCg0KQU1FcyBNb2RlbCA0IChubyBTRXMpDQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQogICAgZGZtYWogPC0gZGZtaW4gPC0gZGZtZW4gPC0gZGZ3b21lbiA8LSBkZl9wcGY1DQogICAgZGZ3b21lbiRnZW5kZXIgPC0gIndvbWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCB3b21lbg0KICAgIGRmbWVuJGdlbmRlciA8LSAibWVuIiAjIG9uZSBkYXRhc2V0IG9mIGFsbCBtZW4NCiAgICBkZm1pbiRldGhuaWNpdHkzIDwtICJtaW5vcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWlub3JpdHkNCiAgICBkZm1haiRldGhuaWNpdHkzIDwtICJtYWpvcml0eSIgIyBvbmUgZGF0YXNldCBhbGwgbWFqb3JpdHkNCiAgICANCiAgICBkZndvbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZ3b21lbiRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRnZW5kZXIpKQ0KICAgIGRmbWVuJGdlbmRlciA8LSBmYWN0b3IoZGZtZW4kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1pbiRldGhuaWNpdHkzIDwtIGZhY3RvcihkZm1pbiRldGhuaWNpdHkzLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZXRobmljaXR5MykpDQogICAgZGZtYWokZXRobmljaXR5MyA8LSBmYWN0b3IoZGZtYWokZXRobmljaXR5MywgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTMpKQ0KDQoNCiAgICAjY2FsY3VsYXRlIHRoZSBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcyBmb3IgZ2VuZGVyDQogICAgcHcgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmd29tZW4pKSkNCiAgICBwbSA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFI3LCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZtZW4pKSkNCiAgICANCiAgICAjIGF2ZXJhZ2UgbWFyZ2luYWwgZWZmZWN0cw0KICAgIEFNRV9nZW5kZXIgPC0gbWVhbihwdyAtIHBtKQ0KICAgIA0KICAgICNjYWxjdWxhdGUgdGhlIHByZWRpY3RlZCBwcm9iYWJpbGl0aWVzIGZvciBldGhuaWNpdHkNCiAgICBwbW4gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWluKSkpDQogICAgcG1qIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjcsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZm1haikpKQ0KDQogICAgIyBhdmVyYWdlIG1hcmdpbmFsIGVmZmVjdHMNCiAgICBBTUVfZXRobmljaXR5IDwtIG1lYW4ocG1uIC0gcG1qKQ0KICAgIA0KICAgIA0KICAgICMgY29ob3J0IGludGVyYWN0aW9ucw0KICAgIA0KICAgIHMgPC0gMC4xDQogICAgIyBjb3B5aW5nIG91ciBkYXRhIGludG8gc2VwYXJhdGUgZGF0YWZyYW1lcw0KICAgIGRmd29tZW5jcCA8LSBkZndvbWVuY20gPC0gZGZtZW5jcCA8LSBkZm1lbmNtIDwtIGRmX3BwZjUNCiAgICBkZm1pbmNwIDwtIGRmbWluY20gPC0gZGZtYWpjcCA8LSBkZm1hamNtIDwtIGRmX3BwZjUNCiAgICANCiAgICAjIGFzc2lnbmluZyBnZW5kZXIgdG8gZWFjaCBvZiB0aGUgZGF0YWZyYW1lcw0KICAgIGRmd29tZW5jcCRnZW5kZXIgPC0gZGZ3b21lbmNtJGdlbmRlciA8LSAid29tZW4iDQogICAgZGZtZW5jcCRnZW5kZXIgPC0gZGZtZW5jbSRnZW5kZXIgPC0gIm1lbiINCiAgICBkZndvbWVuY3AkZ2VuZGVyIDwtIGZhY3RvcihkZndvbWVuY3AkZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZndvbWVuY20kZ2VuZGVyIDwtIGZhY3RvcihkZndvbWVuY20kZ2VuZGVyLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZ2VuZGVyKSkNCiAgICBkZm1lbmNwJGdlbmRlciA8LSBmYWN0b3IoZGZtZW5jcCRnZW5kZXIsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRnZW5kZXIpKQ0KICAgIGRmbWVuY20kZ2VuZGVyIDwtIGZhY3RvcihkZm1lbmNtJGdlbmRlciwgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGdlbmRlcikpDQogICAgDQogICAgIyBhc3NpZ25pbmcgZXRobmljaXR5IHRvIHRoZSBkYXRhZnJhbWVzDQogICAgZGZtaW5jcCRldGhuaWNpdHkzIDwtIGRmbWluY20kZXRobmljaXR5MyA8LSAibWlub3JpdHkiDQogICAgZGZtYWpjcCRldGhuaWNpdHkzIDwtIGRmbWFqY20kZXRobmljaXR5MyA8LSAibWFqb3JpdHkiDQogICAgZGZtaW5jcCRldGhuaWNpdHkzIDwtIGZhY3RvcihkZm1pbmNwJGV0aG5pY2l0eTMsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRldGhuaWNpdHkzKSkNCiAgICBkZm1pbmNtJGV0aG5pY2l0eTMgPC0gZmFjdG9yKGRmbWluY20kZXRobmljaXR5MywgbGV2ZWxzPWxldmVscyhkZl9wcGY1JGV0aG5pY2l0eTMpKQ0KICAgIGRmbWFqY3AkZXRobmljaXR5MyA8LSBmYWN0b3IoZGZtYWpjcCRldGhuaWNpdHkzLCBsZXZlbHM9bGV2ZWxzKGRmX3BwZjUkZXRobmljaXR5MykpDQogICAgZGZtYWpjbSRldGhuaWNpdHkzIDwtIGZhY3RvcihkZm1hamNtJGV0aG5pY2l0eTMsIGxldmVscz1sZXZlbHMoZGZfcHBmNSRldGhuaWNpdHkzKSkNCiAgICANCiAgICANCiAgICAjIGFkZGluZy9zdWJ0cmFjdGluZyBzbWFsbCBjaGFuZ2VzIFBoRCBjb2hvcnQNCiAgICBkZndvbWVuY3AkcGhkX2NvaG9ydCA8LSBkZm1lbmNwJHBoZF9jb2hvcnQgPC0gZGZfcHBmNSRwaGRfY29ob3J0ICsgcw0KICAgIGRmd29tZW5jbSRwaGRfY29ob3J0IDwtIGRmbWVuY20kcGhkX2NvaG9ydCA8LSBkZl9wcGY1JHBoZF9jb2hvcnQgLSBzDQogICAgDQogICAgZGZtYWpjcCRwaGRfY29ob3J0IDwtIGRmbWluY3AkcGhkX2NvaG9ydCA8LSBkZl9wcGY1JHBoZF9jb2hvcnQgKyBzDQogICAgZGZtYWpjbSRwaGRfY29ob3J0IDwtIGRmbWluY20kcGhkX2NvaG9ydCA8LSBkZl9wcGY1JHBoZF9jb2hvcnQgLSBzDQogICAgDQogICAgIyBjYWxjdWxhdGluZyBwcmVkaWN0ZWQgcHJvYmFiaWxpdGllcw0KICAgIHB3cCA8LSBhcy5udW1lcmljKHVubGlzdChwcmVkaWN0KFI3LCB0eXBlPSJyZXNwb25zZSIsIG5ld2RhdGE9ZGZ3b21lbmNwKSkpDQogICAgcHdtIDwtIGFzLm51bWVyaWModW5saXN0KHByZWRpY3QoUjcsIHR5cGU9InJlc3BvbnNlIiwgbmV3ZGF0YT1kZndvbWVuY20pKSkNCiAgICBwbXAgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuY3ApKSkNCiAgICBwbW0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWVuY20pKSkNCiAgICANCiAgICBwbnAgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWluY3ApKSkNCiAgICBwbm0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWluY20pKSkNCiAgICBwanAgPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWFqY3ApKSkNCiAgICBwam0gPC0gYXMubnVtZXJpYyh1bmxpc3QocHJlZGljdChSNywgdHlwZT0icmVzcG9uc2UiLCBuZXdkYXRhPWRmbWFqY20pKSkNCiAgICANCiAgICANCiMgbWFyZ2luYWwgZWZmZWN0cw0KQU1FX2dlbmRlcmNvaCA8LSBtZWFuKCgocHdwIC0gcG1wKSAtIChwd20gLSBwbW0pKSAvICgyKnMpKQ0KDQpBTUVfZXRobmljaXR5Y29oIDwtIG1lYW4oKChwbnAgLSBwanApIC0gKHBubSAtIHBqbSkpIC8gKDIqcykpDQoNCg0KQU1FX2dlbmRlcg0KQU1FX2dlbmRlcmNvaA0KDQpBTUVfZXRobmljaXR5DQpBTUVfZXRobmljaXR5Y29oDQoNCmBgYA0KDQoNCiMjIFIzOiBkaXNjcmV0ZS10aW1lIGV2ZW50IGhpc3RvcnkgcmVncmVzc2lvbg0KDQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQpkZl9wcGYzICU+JQ0KICBncm91cF9ieSh0aW1lKSAlPiUNCiAgc3VtbWFyaXNlKGV2ZW50ID0gc3VtKGluYWN0aXZlKSwNCiAgICAgICAgICAgIHRvdGFsID0gbigpKSAlPiUNCiAgbXV0YXRlKGhhemFyZCA9IGV2ZW50L3RvdGFsKSAlPiUNCiAgZ2dwbG90KGFlcyh4PXRpbWUsIHkgPSBsb2coLWxvZygxLWhhemFyZCkpKSkgKw0KICBnZW9tX3BvaW50KCkgKw0KICBnZW9tX3Ntb290aCgpDQoNCiMgQy1sb2ctbG9nIGxpbmsNCg0KYGBgDQoNCg0KYGBge3IsIGV2YWw9RkFMU0V9DQoNCg0KUjggPC0gZ2xtKGZvcm11bGEgPSBpbmFjdGl2ZSB+IHRpbWUgKyBnZW5kZXIsDQogICAgICAgICAgZmFtaWx5ID0gYmlub21pYWwobGluaz0iY2xvZ2xvZyIpLA0KICAgICAgICAgIGRhdGEgPSBkZl9wcGYzKQ0KDQpSOSA8LSBnbG0oZm9ybXVsYSA9IGluYWN0aXZlIH4gdGltZSArIGV0aG5pY2l0eTIsDQogICAgICAgICAgZmFtaWx5ID0gYmlub21pYWwobGluaz0iY2xvZ2xvZyIpLA0KICAgICAgICAgIGRhdGEgPSBkZl9wcGYzKQ0KDQpSMTAgPC0gZ2xtKGZvcm11bGEgPSBpbmFjdGl2ZSB+IHRpbWUgKyBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQgKyBwaGRfY29ob3J0ICsgbnB1YnNfcHJldl9zLA0KICAgICAgICAgIGZhbWlseSA9IGJpbm9taWFsKGxpbms9ImNsb2dsb2ciKSwNCiAgICAgICAgICBkYXRhID0gZGZfcHBmMykNCg0KUjExIDwtIGdsbShmb3JtdWxhID0gaW5hY3RpdmUgfiB0aW1lICsgZ2VuZGVyICsgZXRobmljaXR5MiArIHVuaSArIGZpZWxkICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcyArIGdlbmRlcipwaGRfY29ob3J0ICsgZXRobmljaXR5MipwaGRfY29ob3J0LA0KICAgICAgICAgIGZhbWlseSA9IGJpbm9taWFsKGxpbms9ImNsb2dsb2ciKSwNCiAgICAgICAgICBkYXRhID0gZGZfcHBmMykNCg0Kc3VtbWFyeShSOCwgZXhwID0gVCkNCnN1bW1hcnkoUjksIGV4cCA9IFQpDQpzdW1tYXJ5KFIxMCwgZXhwID0gVCkNCnN1bW1hcnkoUjExLCBleHAgPSBUKQ0KDQoNCmBgYA0KDQoNCg0KIyMgUjQ6IHJvYnVzdG5lc3Mgb2YgZ2VuZGVyLWNvaG9ydCBpbnRlcmFjdGlvbiB0byBjaG9pY2Ugb2YgY29ob3J0cw0KDQpJbiBjb2hvcnQgMyAoaS5lLiAxOTkzKSwgRmlndXJlIDYgc2hvd3MgYSBkaXZlcmdlbnQgZ2VuZGVyIHBhdHRlcm46IHdvbWVuIHRlbmQgdG8gc3Vydml2ZSBtdWNoIGxvbmdlciB0aGFuIG1lbiBpbiB0aGlzIHNwZWNpZmljIGNvaG9ydC4gSGVuY2UsIHRoZSBvYnNlcnZlZCBjb2hvcnQgZWZmZWN0IG1pZ2h0IGJlIGR1ZSB0byB0aGlzIGV4Y2VwdGlvbmFsIHllYXIuIFdlIGxvb2sgaW50byB0aGlzIGJ5IHJlbW92aW5nIHRoaXMgY29ob3J0IGFuZCByZXJ1bm5pbmcgbW9kZWwgNC4gDQoNCmBgYHtyLCBldmFsPUZBTFNFfQ0KDQojIFJlbW92aW5nIGNvaG9ydCAzDQpkZl9ub2NvaDIgPC0gZGZfcHBmM1tkZl9wcGYzJHBoZF9jb2hvcnQhPTIsXQ0KDQpNNF9ubzIgPC0gZmxleHN1cnZyZWcoU3VydigodGltZSAtMSksIHRpbWUsIGluYWN0aXZlKSB+IGdlbmRlciArIGV0aG5pY2l0eTIgKyB1bmkgKyBmaWVsZDIgKyBwaGRfY29ob3J0ICsgbnB1YnNfcHJldl9zICsgcGhkX2NvaG9ydCpnZW5kZXIgKyBwaGRfY29ob3J0KmV0aG5pY2l0eTIsIGRhdGEgPSBkZl9ub2NvaDIsIGRpc3QgPSAibG9nbm9ybWFsIikNCg0KTTRfbm8yDQoNCg0KIyBSZW1vdmluZyBjb2hvcnQgMC0yDQpkZl9ub2NvaDAyIDwtIGRmX3BwZjNbZGZfcHBmMyRwaGRfY29ob3J0PjIsXQ0KDQpNNF9ubzAyIDwtIGZsZXhzdXJ2cmVnKFN1cnYoKHRpbWUgLTEpLCB0aW1lLCBpbmFjdGl2ZSkgfiBnZW5kZXIgKyBldGhuaWNpdHkyICsgdW5pICsgZmllbGQyICsgcGhkX2NvaG9ydCArIG5wdWJzX3ByZXZfcyArIHBoZF9jb2hvcnQqZ2VuZGVyICsgcGhkX2NvaG9ydCpldGhuaWNpdHkyLCBkYXRhID0gZGZfbm9jb2gwMiwgZGlzdCA9ICJsb2dub3JtYWwiKQ0KDQpNNF9ubzAyDQoNCiMgUmVtb3ZpbmcgY29ob3J0IDAtNA0KZGZfbm9jb2gwNCA8LSBkZl9wcGYzW2RmX3BwZjMkcGhkX2NvaG9ydD40LF0NCg0KTTRfbm8wNCA8LSBmbGV4c3VydnJlZyhTdXJ2KCh0aW1lIC0xKSwgdGltZSwgaW5hY3RpdmUpIH4gZ2VuZGVyICsgZXRobmljaXR5MiArIHVuaSArIGZpZWxkMiArIHBoZF9jb2hvcnQgKyBucHVic19wcmV2X3MgKyBwaGRfY29ob3J0KmdlbmRlciArIHBoZF9jb2hvcnQqZXRobmljaXR5MiwgZGF0YSA9IGRmX25vY29oMDQsIGRpc3QgPSAibG9nbm9ybWFsIikNCg0KTTRfbm8wNA0KDQpgYGANCg0K
Copyright © 2023